Revana Analysis Report
2022-05-04
Revana was developed by
Elias Ulrich, Stefan Pfister and Natalie Jaeger
Part I: Input data
Analysed samples Input data
- ICGC_MB107
- ICGC_MB112
- ICGC_MB113
- ICGC_MB117
- ICGC_MB122
- ICGC_MB124
- ICGC_MB128
- ICGC_MB130
- ICGC_MB140
- ICGC_MB148
- ICGC_MB176
- ICGC_MB31
- ICGC_MB46
- ICGC_MB54
- ICGC_MB20
- ICGC_MB1
- ICGC_MB101
- ICGC_MB102
- ICGC_MB104
- ICGC_MB12
- ICGC_MB132
- ICGC_MB136
- ICGC_MB137
- ICGC_MB178
- ICGC_MB179
- ICGC_MB181
- ICGC_MB193
- ICGC_MB194
- ICGC_MB204
- ICGC_MB206
- ICGC_MB234
- ICGC_MB239
- ICGC_MB243
- ICGC_MB244
- ICGC_MB246
- ICGC_MB250
- ICGC_MB28
- ICGC_MB34
- ICGC_MB35
- ICGC_MB37
- ICGC_MB53
- ICGC_MB56
- ICGC_MB59
- ICGC_MB60
- ICGC_MB61
- ICGC_MB62
- ICGC_MB63
- ICGC_MB68
- ICGC_MB88
- ICGC_MB23
- ICGC_MB106
- ICGC_MB111
- ICGC_MB131
- ICGC_MB134
- ICGC_MB139
- ICGC_MB146
- ICGC_MB16
- ICGC_MB164
- ICGC_MB166
- ICGC_MB170
- ICGC_MB18
- ICGC_MB188
- ICGC_MB198
- ICGC_MB205
- ICGC_MB226
- ICGC_MB235
- ICGC_MB247
- ICGC_MB248
- ICGC_MB39
- ICGC_MB45
- ICGC_MB5
- ICGC_MB50
- ICGC_MB57
- ICGC_MB89
- ICGC_MB9
- ICGC_MB90
- ICGC_MB95
- ICGC_MB96
- ICGC_MB98
- ICGC_MB36
- ICGC_MB108
- ICGC_MB110
- ICGC_MB114
- ICGC_MB115
- ICGC_MB118
- ICGC_MB119
- ICGC_MB121
- ICGC_MB129
- ICGC_MB15
- ICGC_MB165
- ICGC_MB174
- ICGC_MB175
- ICGC_MB177
- ICGC_MB183
- ICGC_MB184
- ICGC_MB185
- ICGC_MB19
- ICGC_MB199
- ICGC_MB2
- ICGC_MB213
- ICGC_MB224
- ICGC_MB227
- ICGC_MB228
- ICGC_MB230
- ICGC_MB232
- ICGC_MB233
- ICGC_MB236
- ICGC_MB237
- ICGC_MB24
- ICGC_MB26
- ICGC_MB38
- ICGC_MB40
- ICGC_MB49
- ICGC_MB58
- ICGC_MB64
- ICGC_MB65
- ICGC_MB7
- ICGC_MB70
- ICGC_MB71
- ICGC_MB83
- ICGC_MB84
- ICGC_MB85
- ICGC_MB86
- ICGC_MB91
- ICGC_MB92
- ICGC_MB94
- ICGC_MB99
- ICGC_MB6
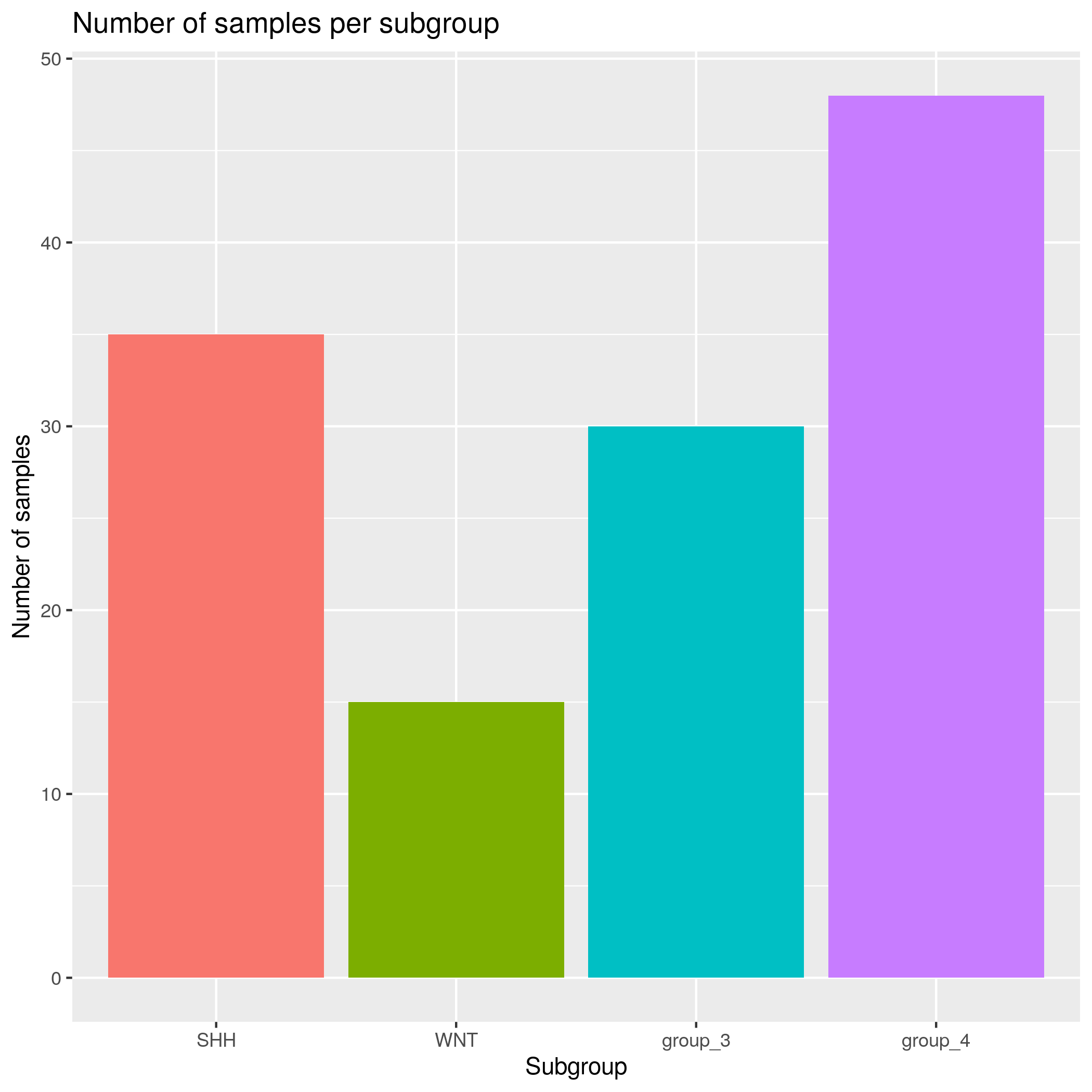
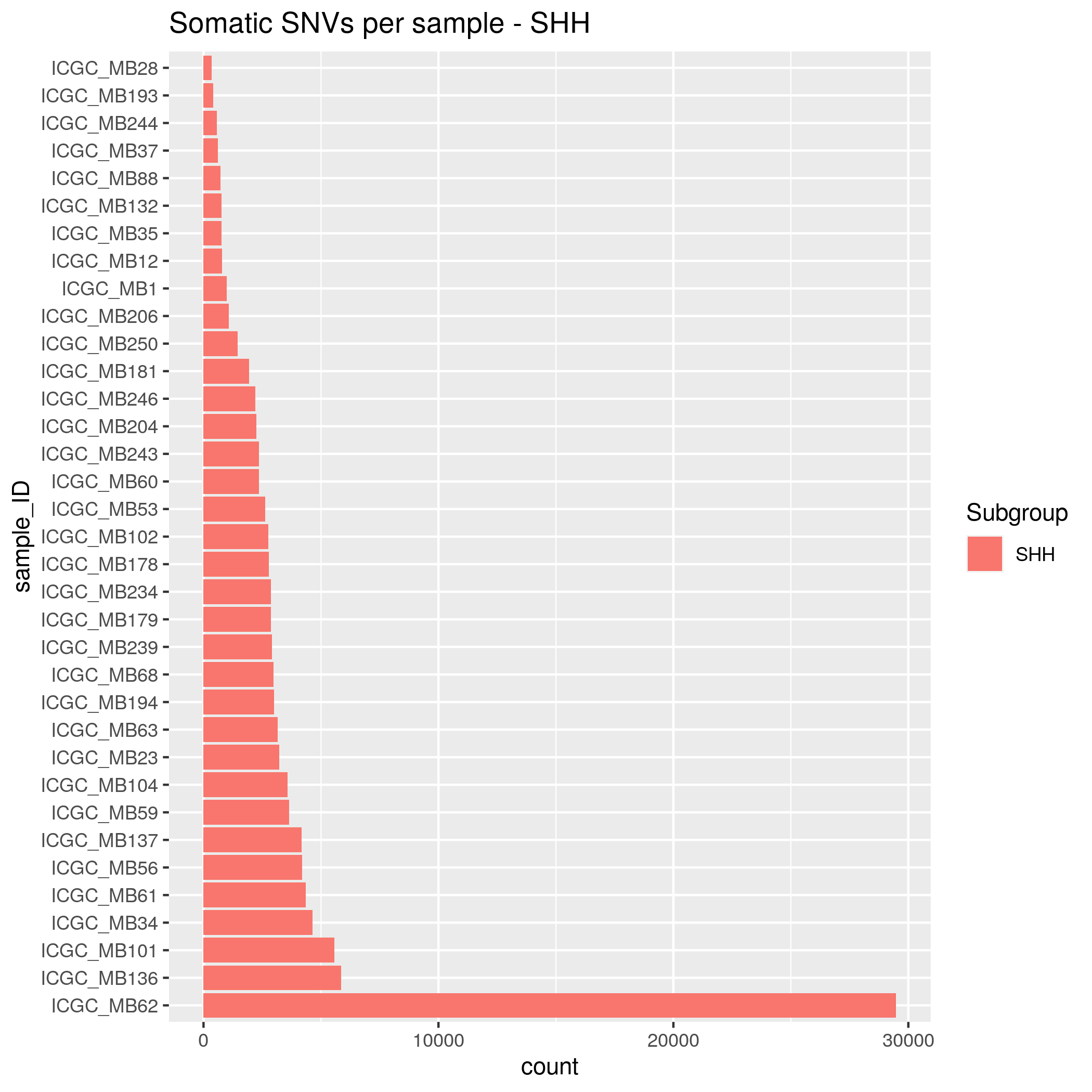
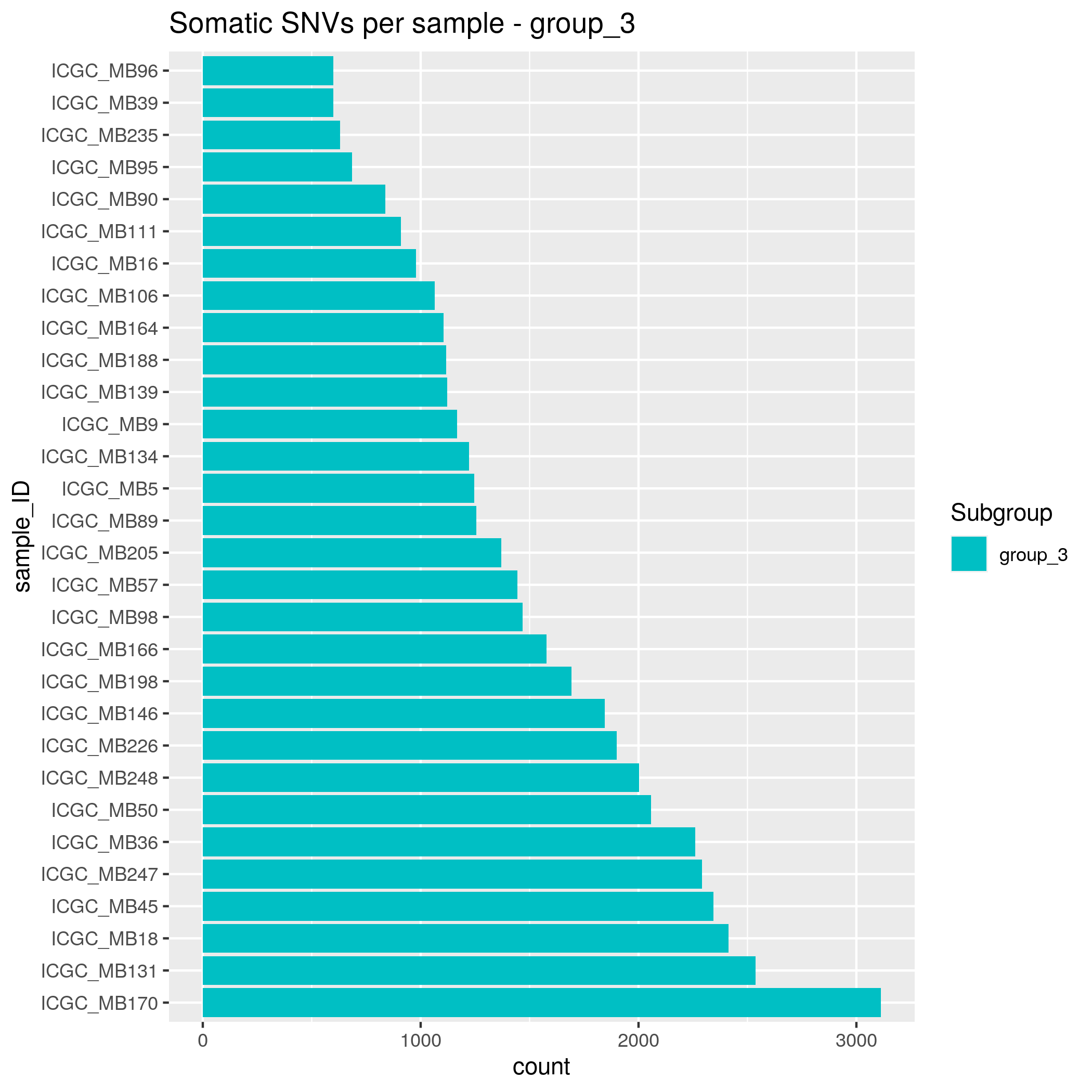
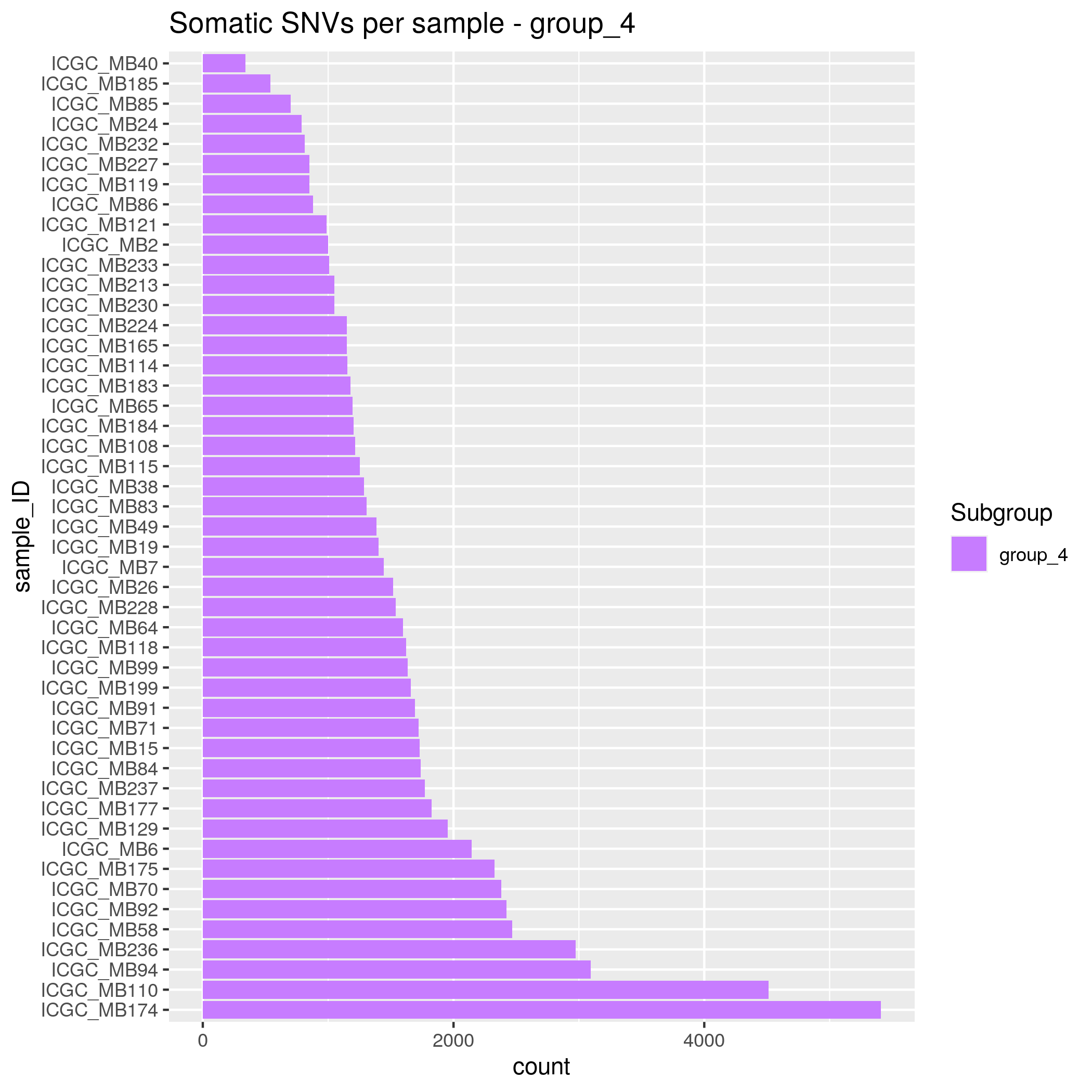
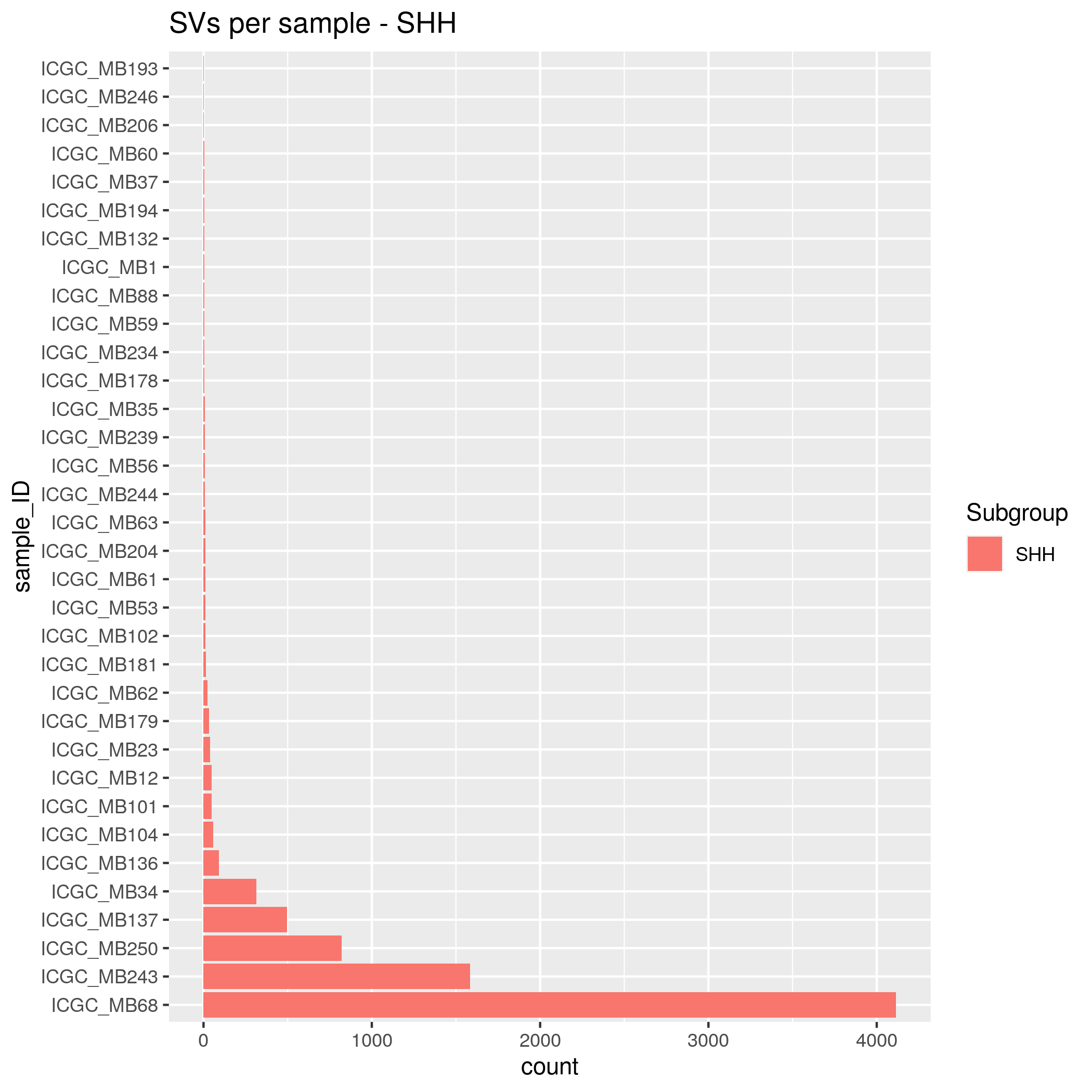
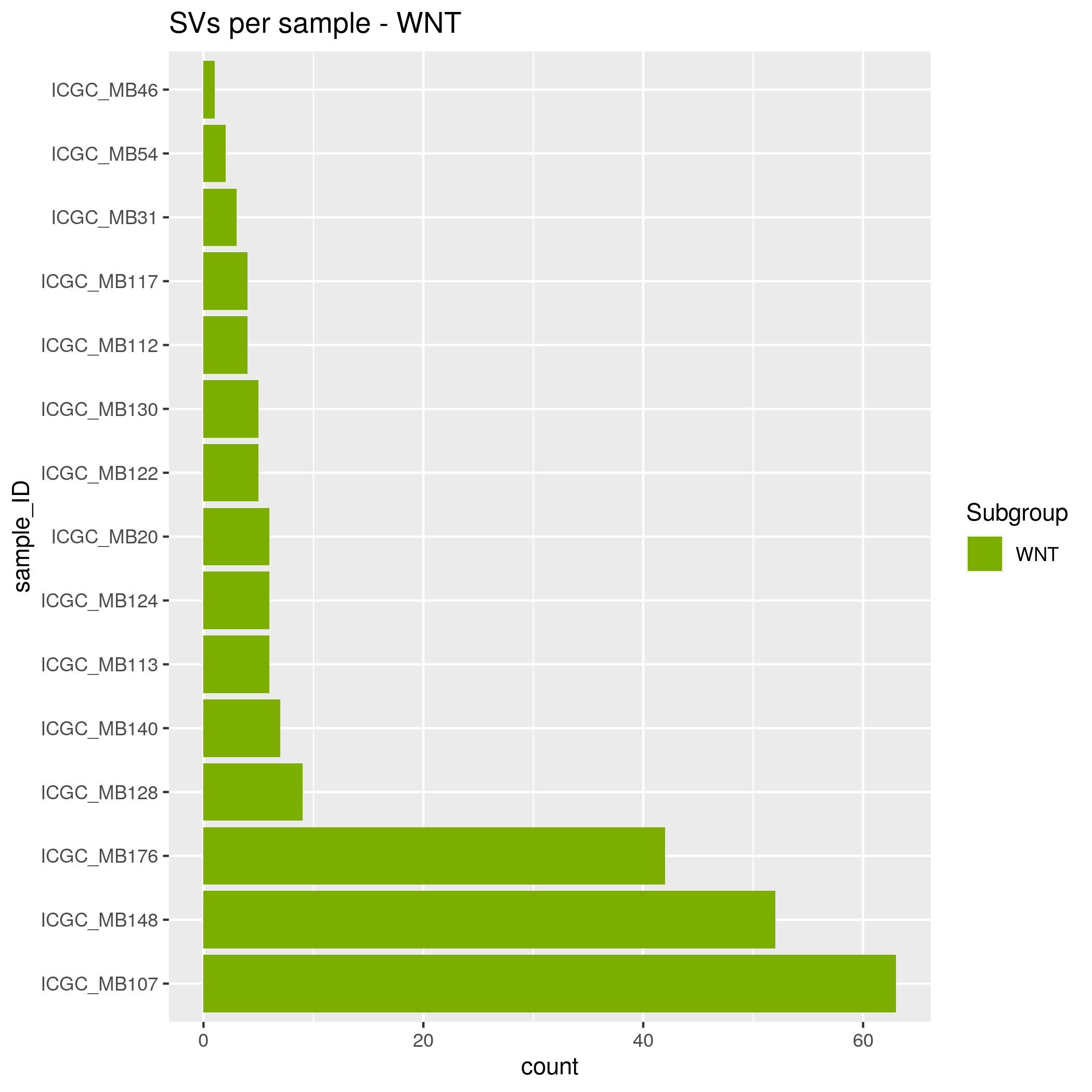
- SHH - 35 samples
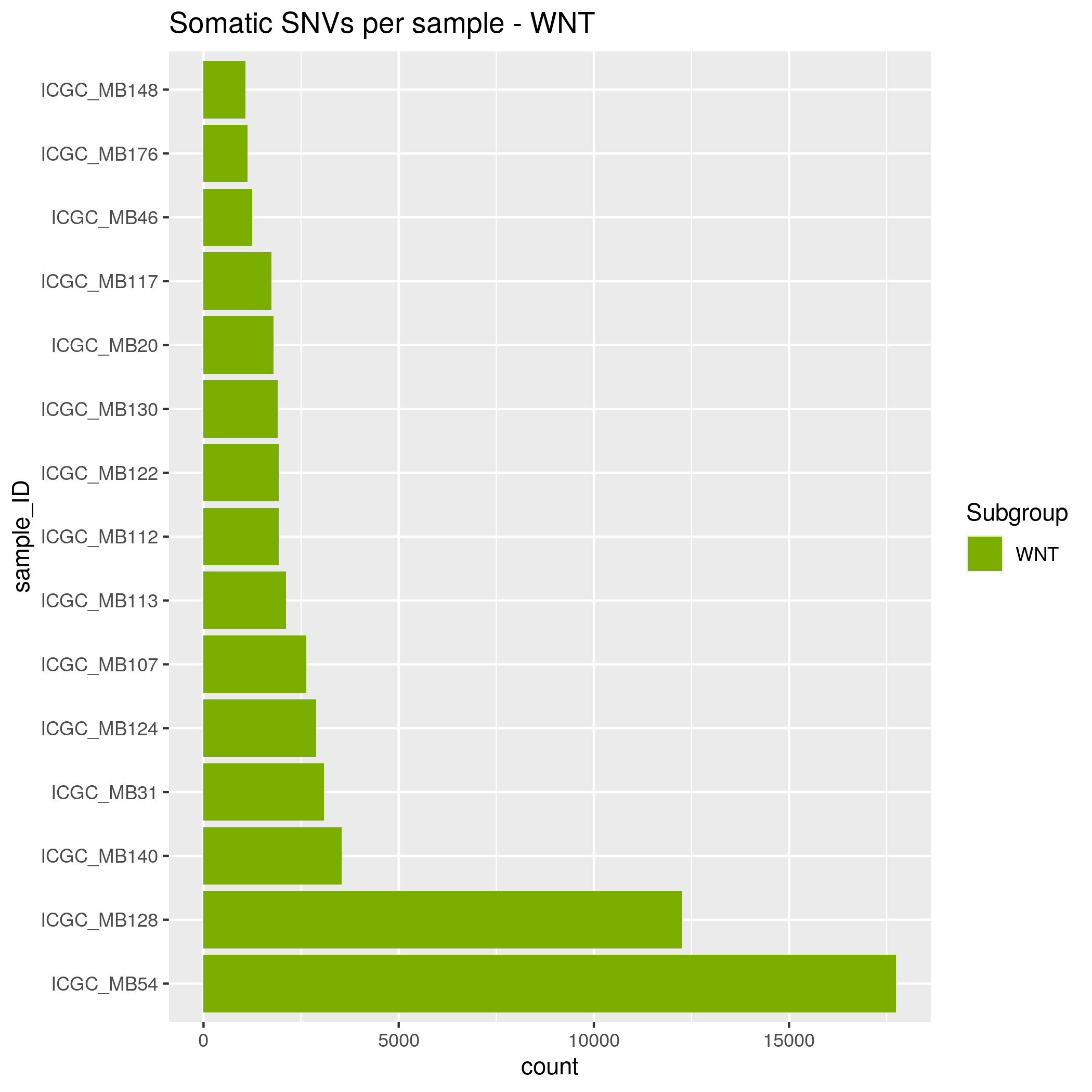
- WNT - 15 samples
- group_3 - 30 samples
- group_4 - 48 samples
Sample Subgroup Composition Input data
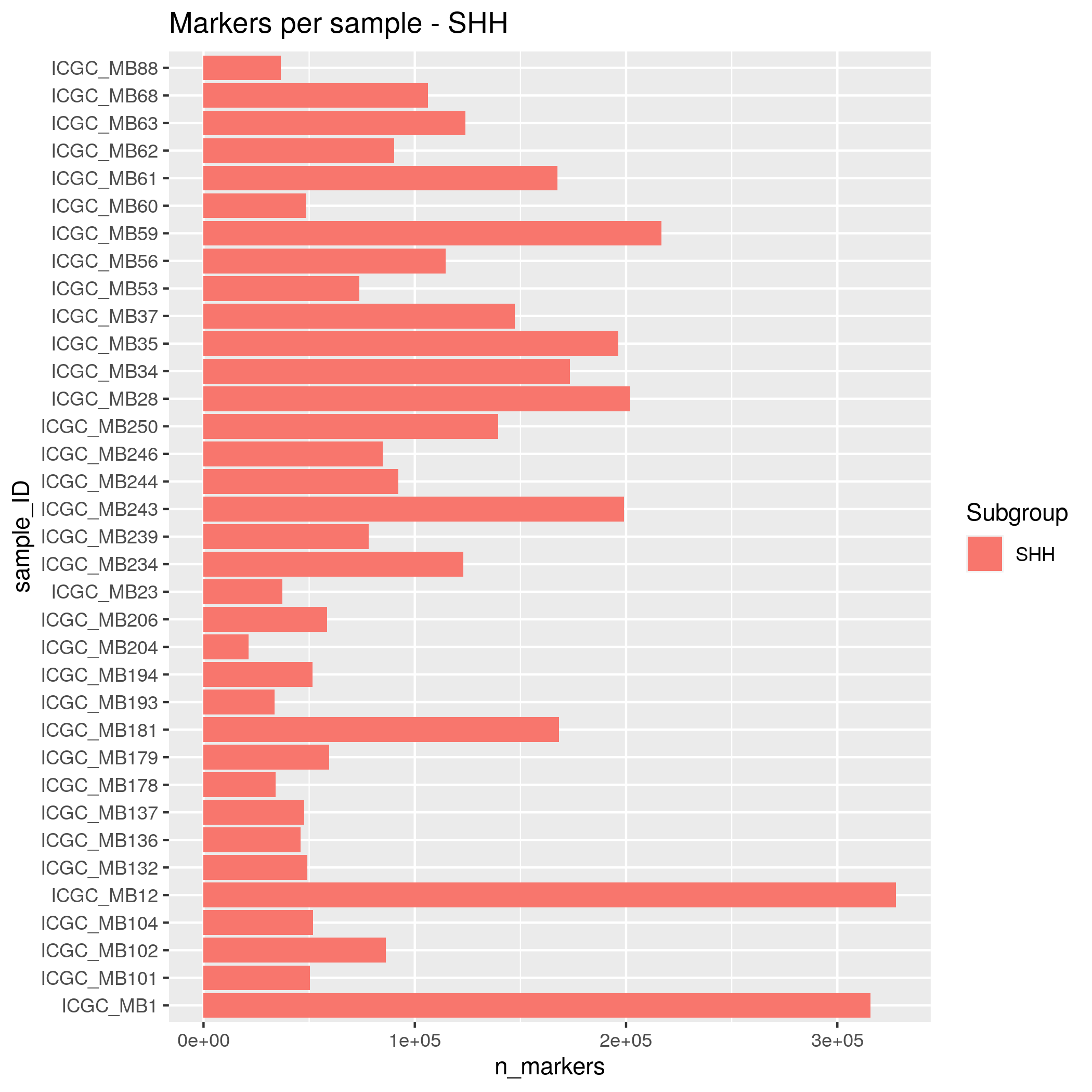
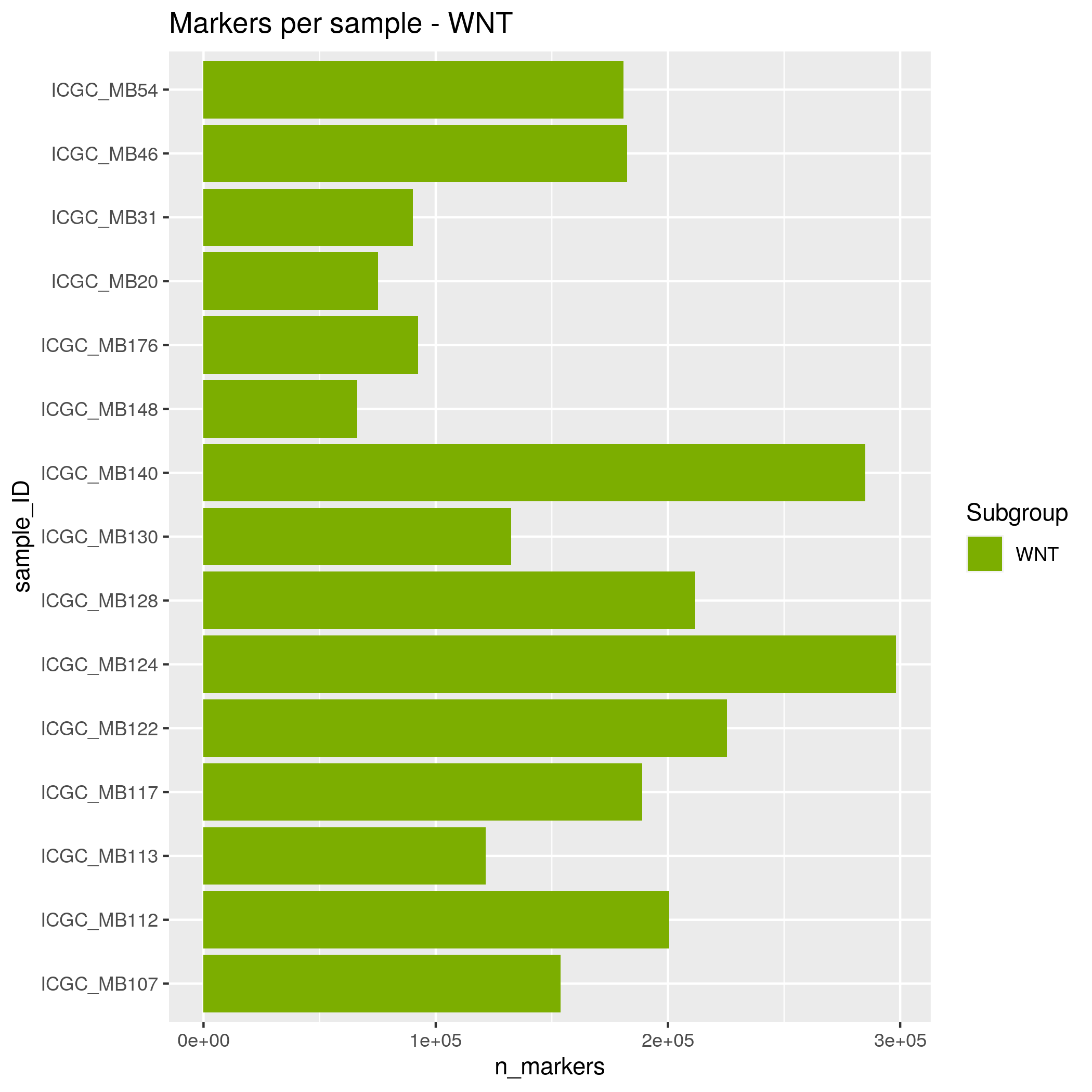
SNP Markers Input data
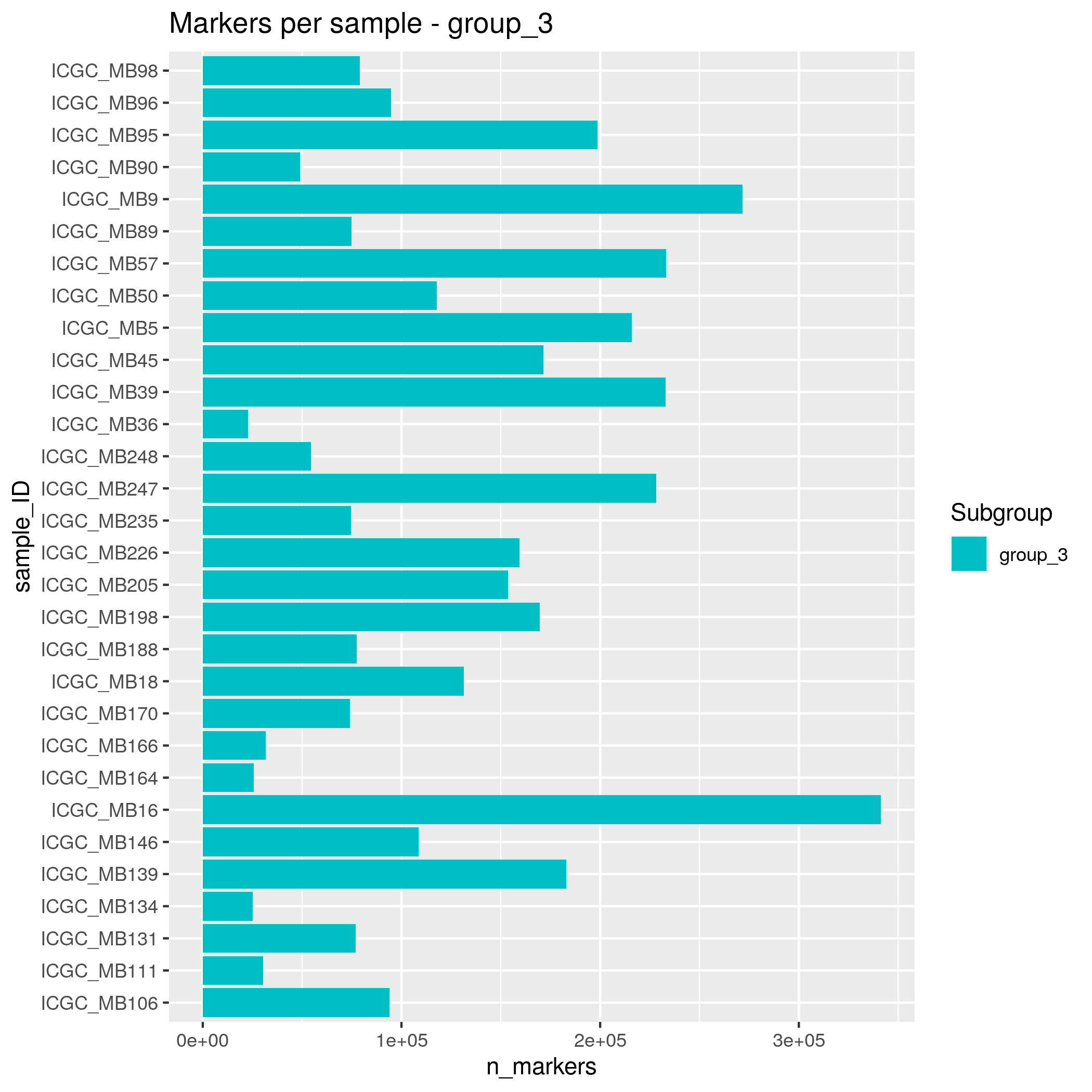
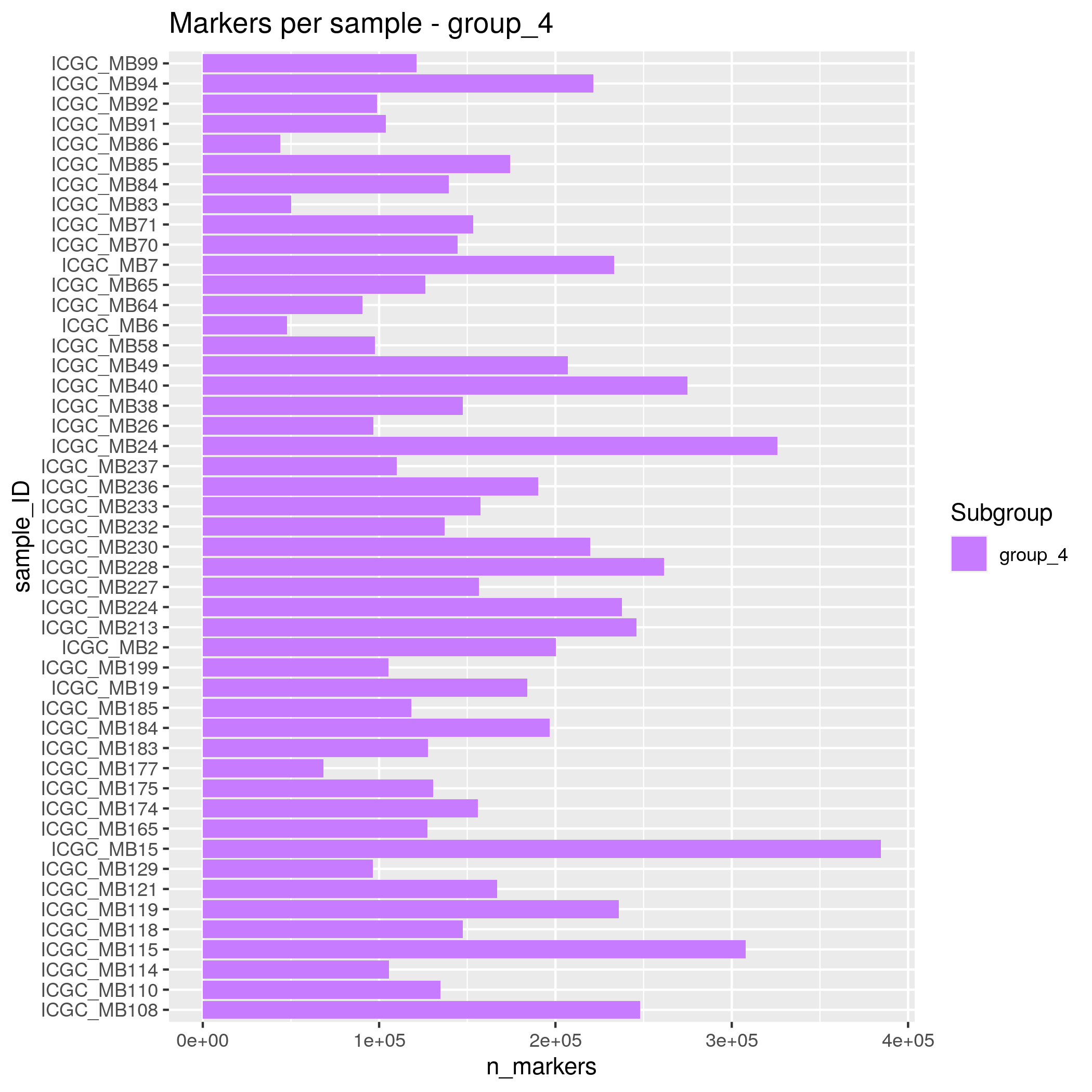
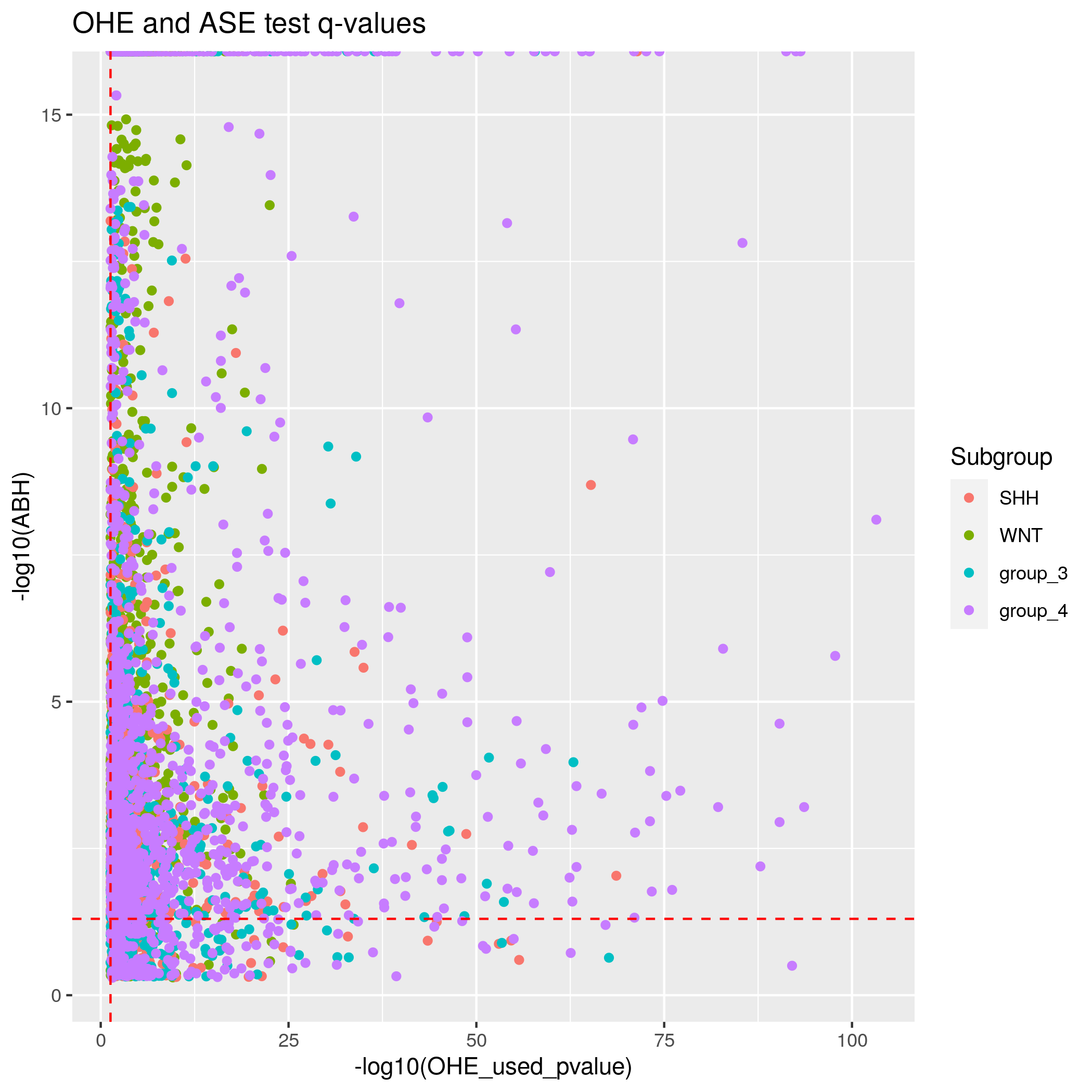
Somatic SNVs/InDels Input data
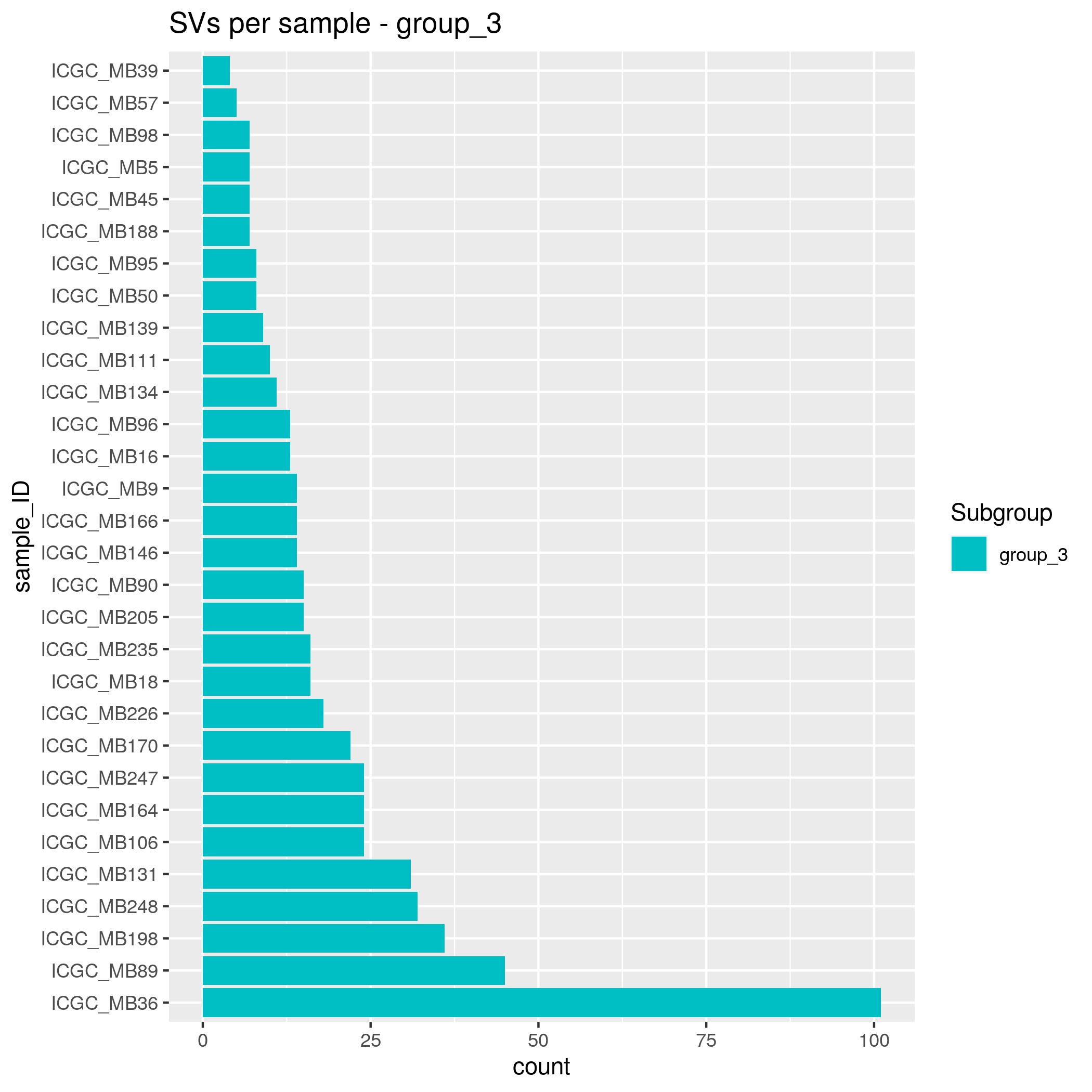
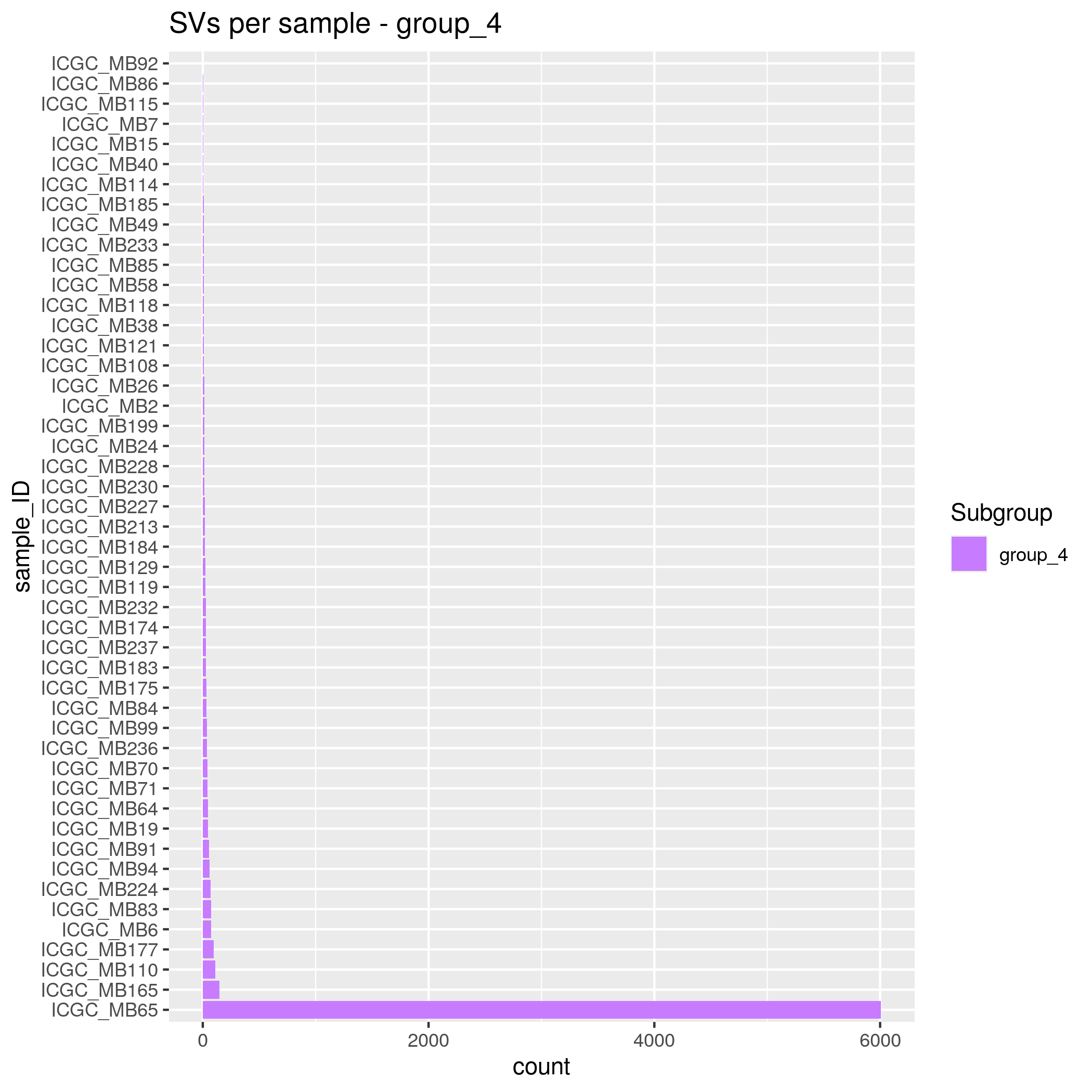
SVs Input data
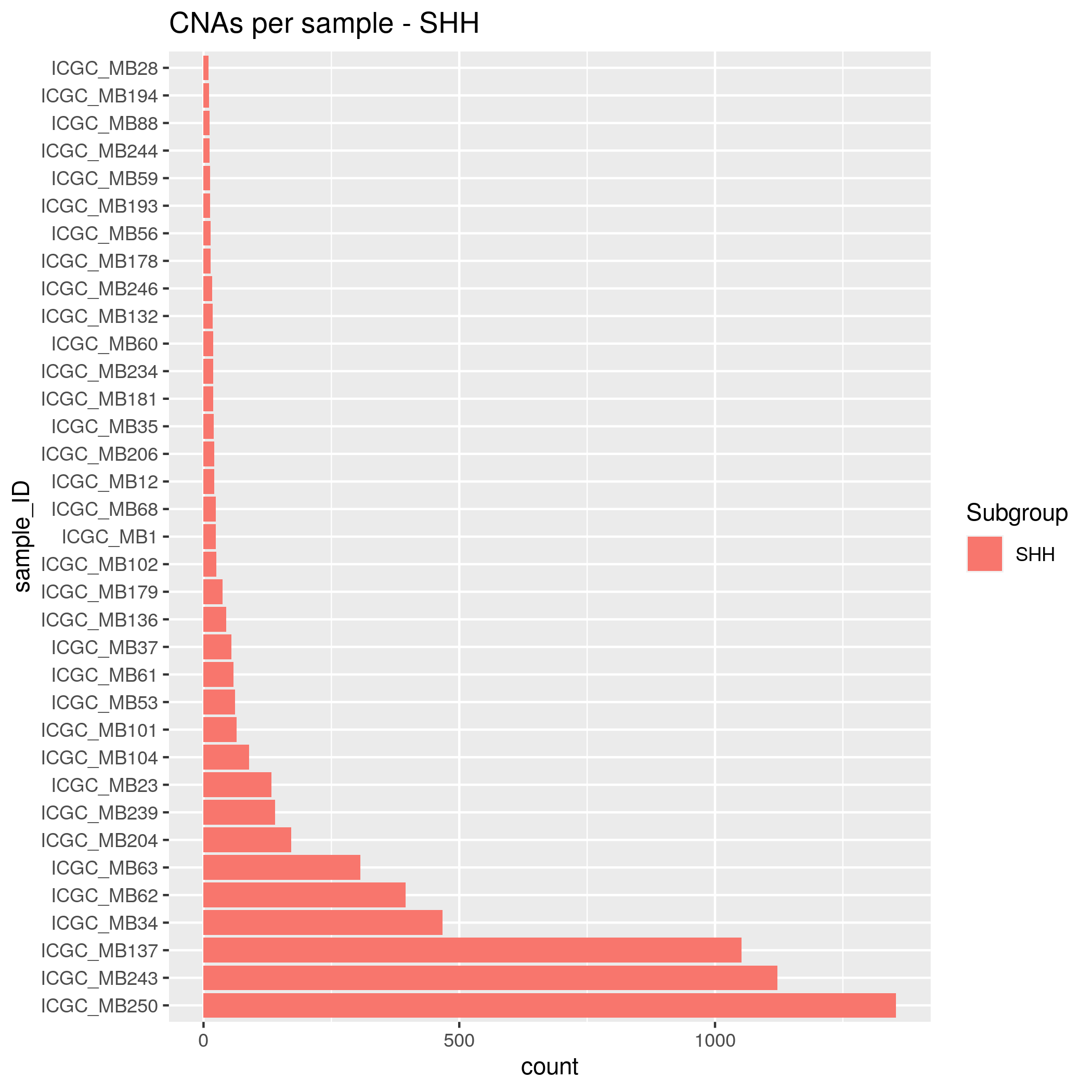
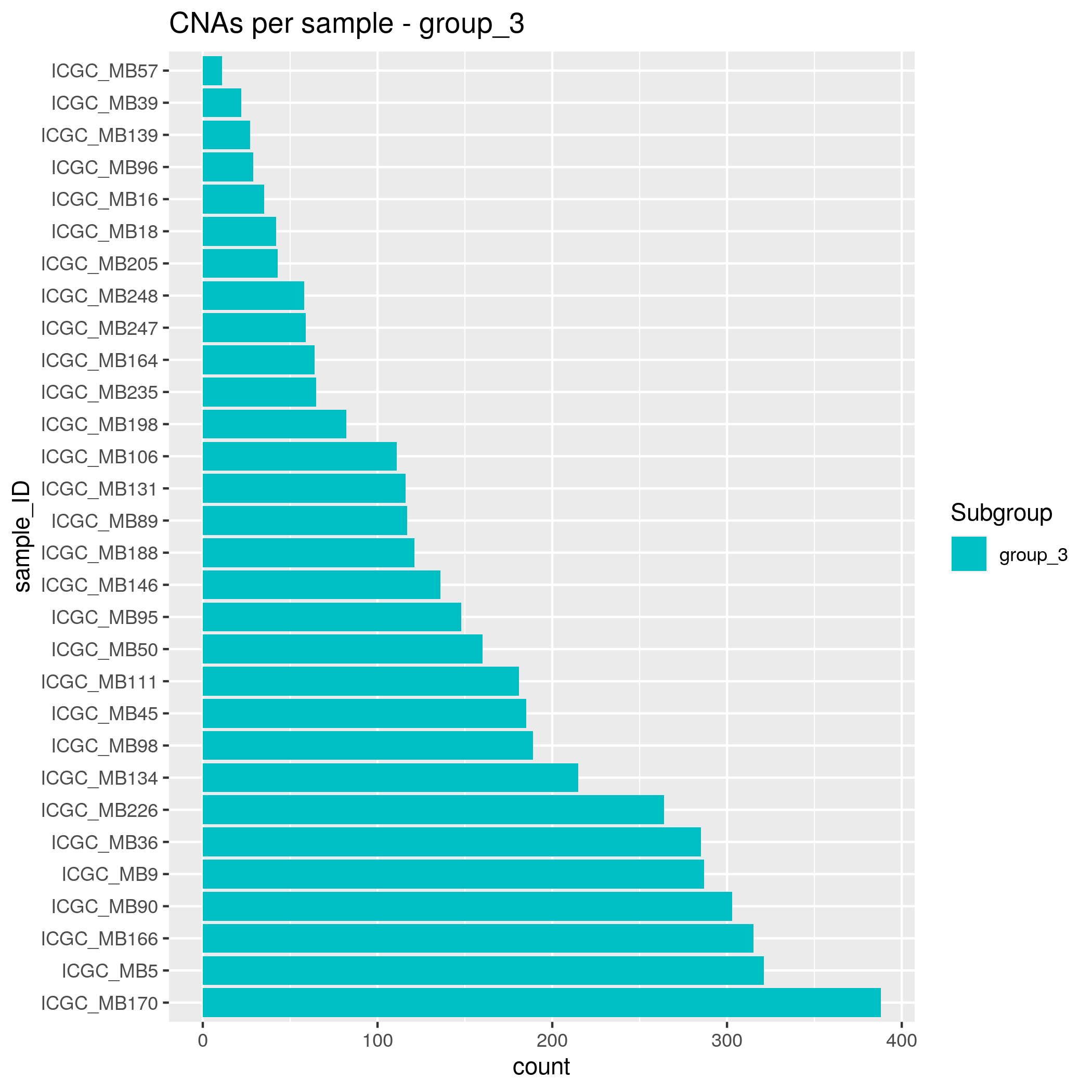
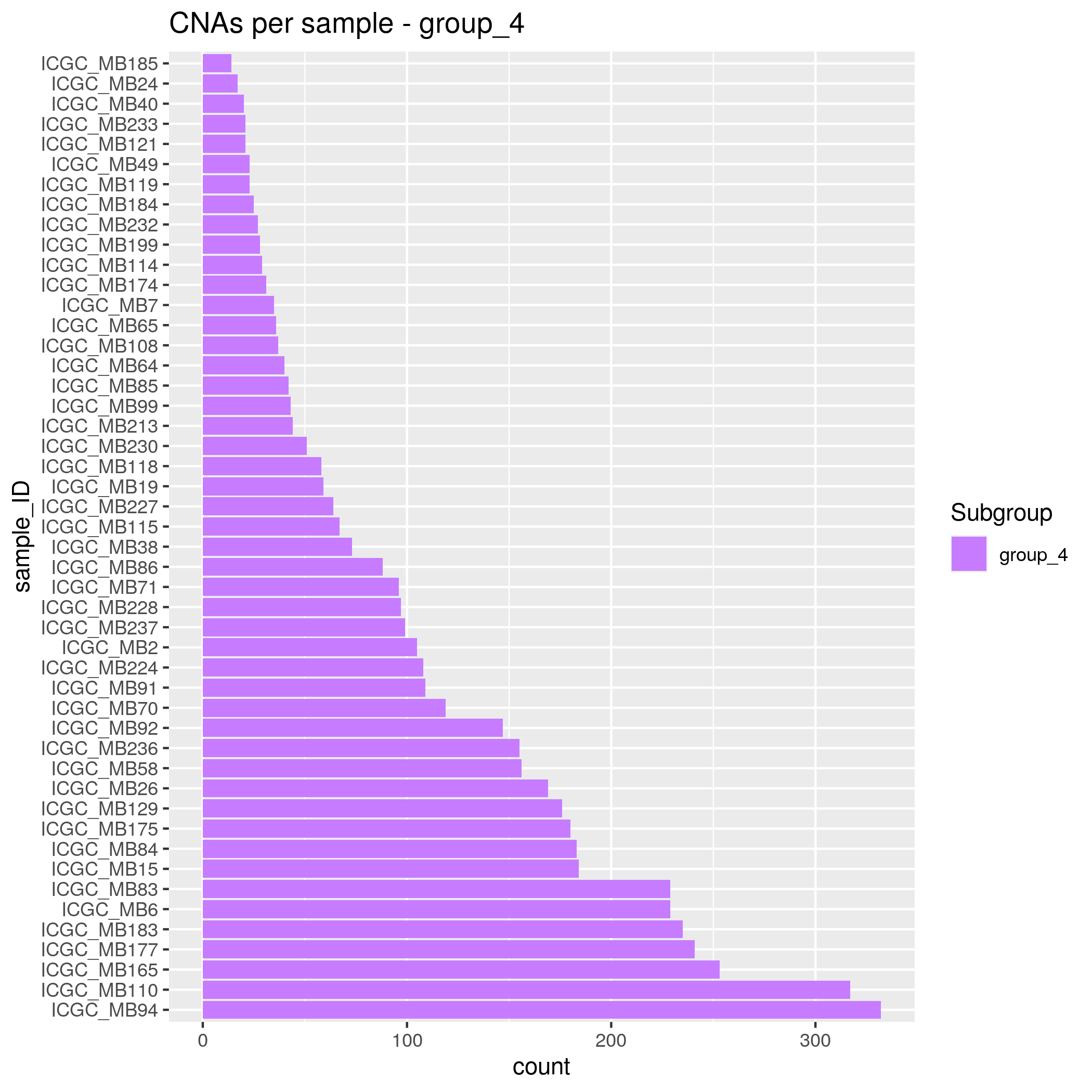
CNAs Input data

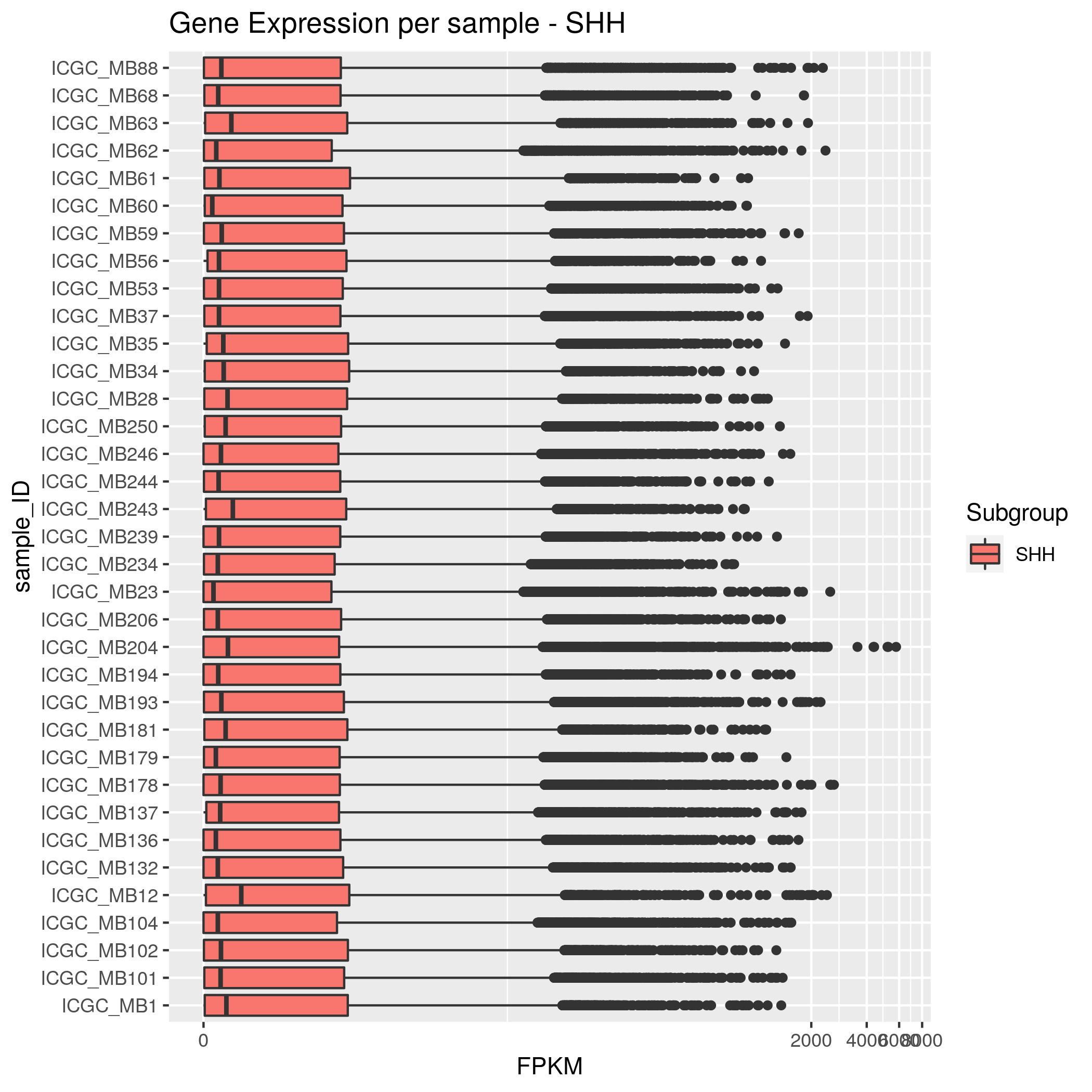
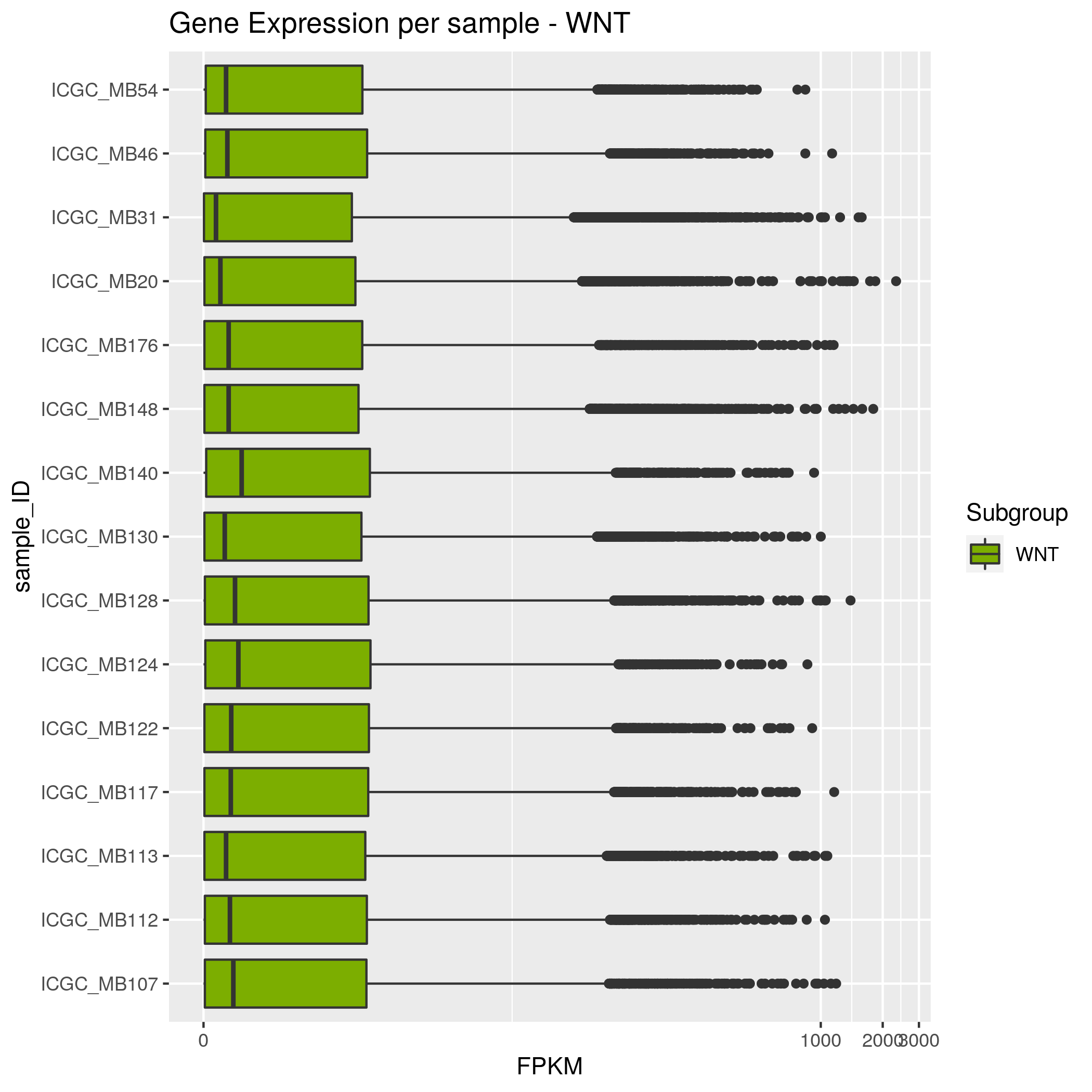
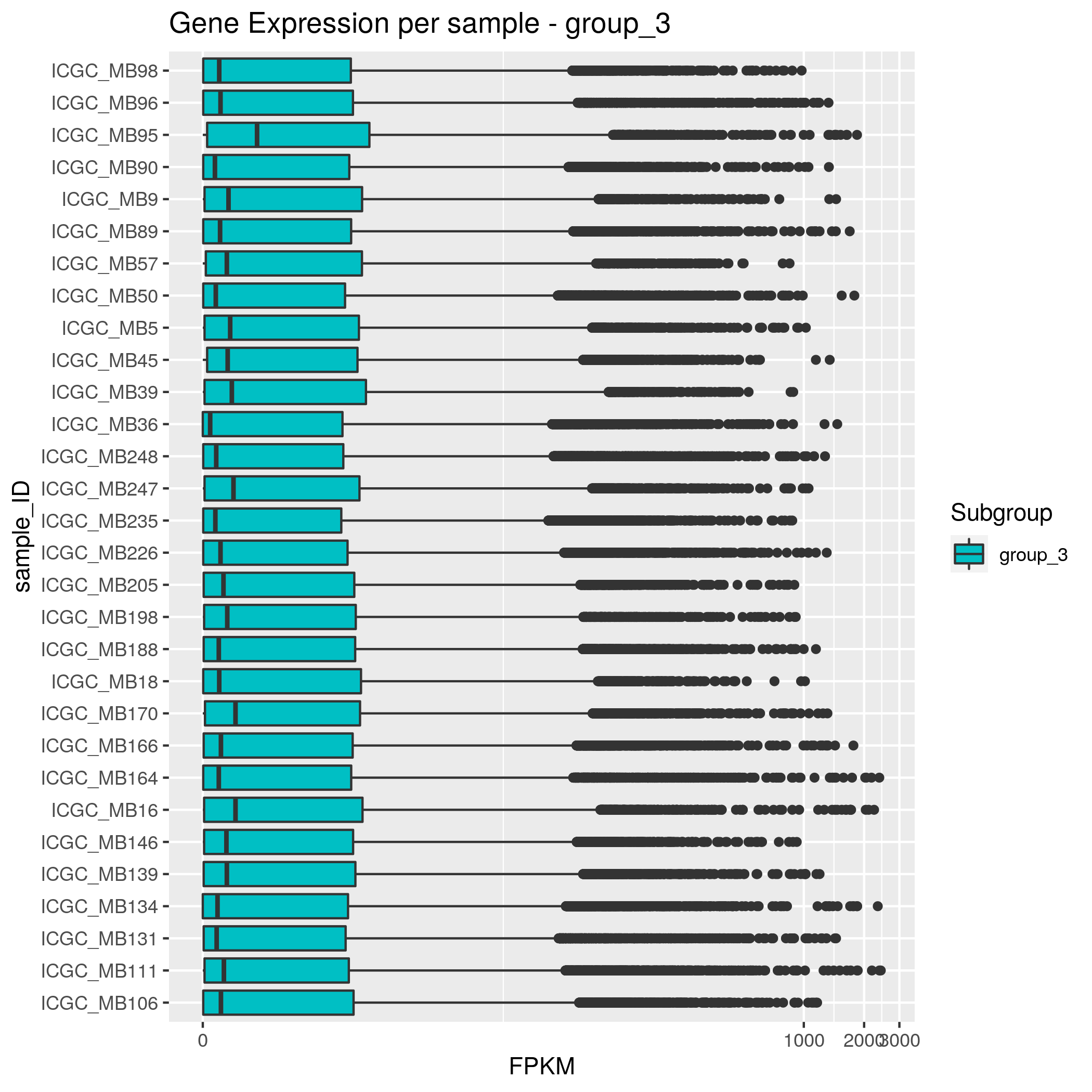
Gene Expression Input data
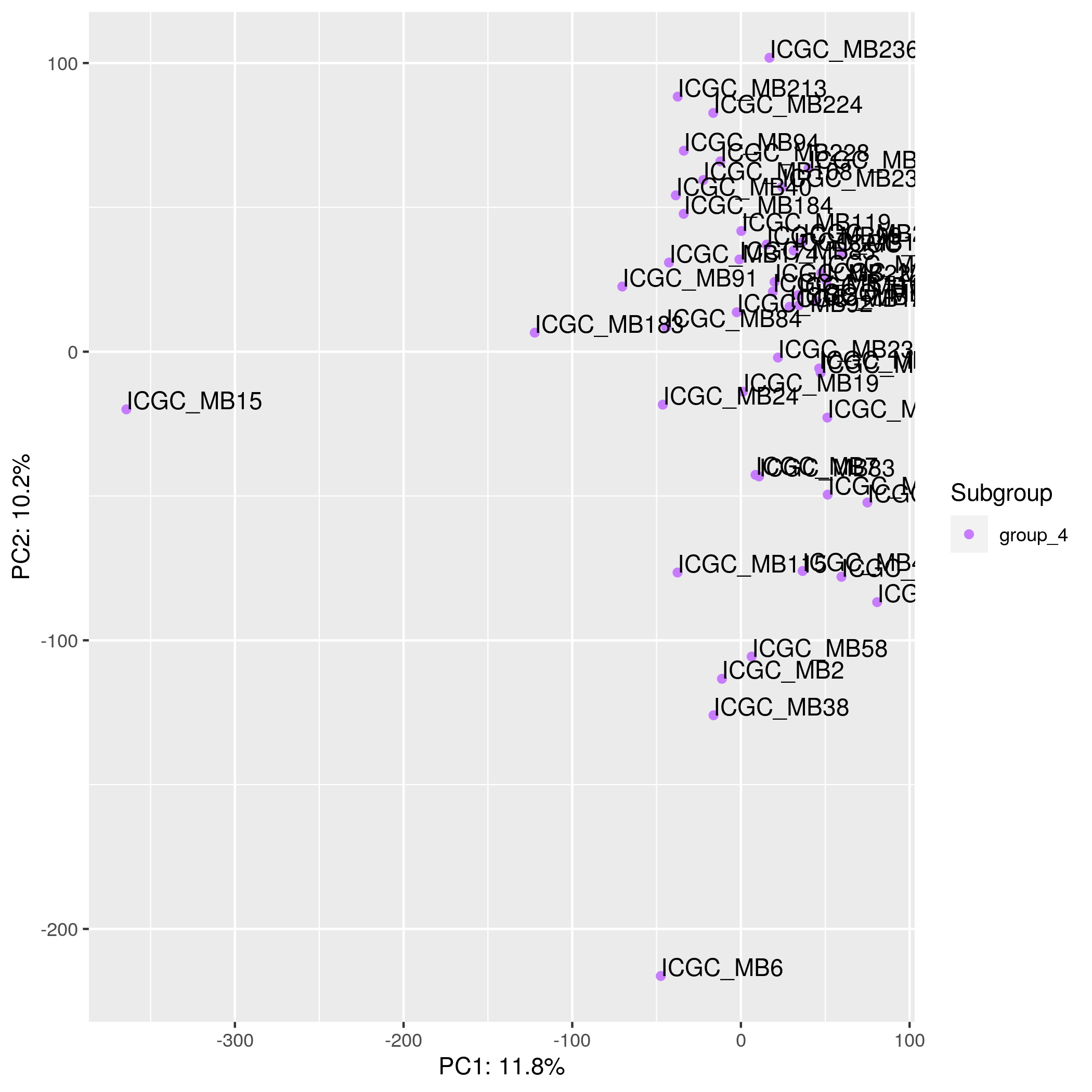
Gene Expression - PCA Input data
Part II: Results Overview
Cis-Activation Overview
- ICGC_MB1
- ICGC_MB101
- ICGC_MB102
- ICGC_MB104
- ICGC_MB106
- ICGC_MB107
- ICGC_MB108
- ICGC_MB110
- ICGC_MB111
- ICGC_MB112
- ICGC_MB113
- ICGC_MB114
- ICGC_MB115
- ICGC_MB117
- ICGC_MB118
- ICGC_MB119
- ICGC_MB12
- ICGC_MB121
- ICGC_MB122
- ICGC_MB124
- ICGC_MB128
- ICGC_MB129
- ICGC_MB130
- ICGC_MB131
- ICGC_MB132
- ICGC_MB134
- ICGC_MB136
- ICGC_MB137
- ICGC_MB139
- ICGC_MB140
- ICGC_MB146
- ICGC_MB148
- ICGC_MB15
- ICGC_MB16
- ICGC_MB164
- ICGC_MB165
- ICGC_MB166
- ICGC_MB170
- ICGC_MB174
- ICGC_MB175
- ICGC_MB176
- ICGC_MB177
- ICGC_MB178
- ICGC_MB179
- ICGC_MB18
- ICGC_MB181
- ICGC_MB183
- ICGC_MB184
- ICGC_MB185
- ICGC_MB188
- ICGC_MB19
- ICGC_MB193
- ICGC_MB194
- ICGC_MB198
- ICGC_MB199
- ICGC_MB2
- ICGC_MB20
- ICGC_MB204
- ICGC_MB205
- ICGC_MB206
- ICGC_MB213
- ICGC_MB224
- ICGC_MB226
- ICGC_MB227
- ICGC_MB228
- ICGC_MB23
- ICGC_MB230
- ICGC_MB232
- ICGC_MB233
- ICGC_MB234
- ICGC_MB235
- ICGC_MB236
- ICGC_MB237
- ICGC_MB239
- ICGC_MB24
- ICGC_MB243
- ICGC_MB244
- ICGC_MB246
- ICGC_MB247
- ICGC_MB248
- ICGC_MB250
- ICGC_MB26
- ICGC_MB28
- ICGC_MB31
- ICGC_MB34
- ICGC_MB35
- ICGC_MB36
- ICGC_MB37
- ICGC_MB38
- ICGC_MB39
- ICGC_MB40
- ICGC_MB45
- ICGC_MB46
- ICGC_MB49
- ICGC_MB5
- ICGC_MB50
- ICGC_MB53
- ICGC_MB54
- ICGC_MB56
- ICGC_MB57
- ICGC_MB58
- ICGC_MB59
- ICGC_MB6
- ICGC_MB60
- ICGC_MB61
- ICGC_MB62
- ICGC_MB63
- ICGC_MB64
- ICGC_MB65
- ICGC_MB68
- ICGC_MB7
- ICGC_MB70
- ICGC_MB71
- ICGC_MB83
- ICGC_MB84
- ICGC_MB85
- ICGC_MB86
- ICGC_MB88
- ICGC_MB89
- ICGC_MB9
- ICGC_MB90
- ICGC_MB91
- ICGC_MB92
- ICGC_MB94
- ICGC_MB95
- ICGC_MB96
- ICGC_MB98
- ICGC_MB99
Cis-activated genes
3952
Unique cis-activated genes
2803
Average cis-activated genes per sample
30.875
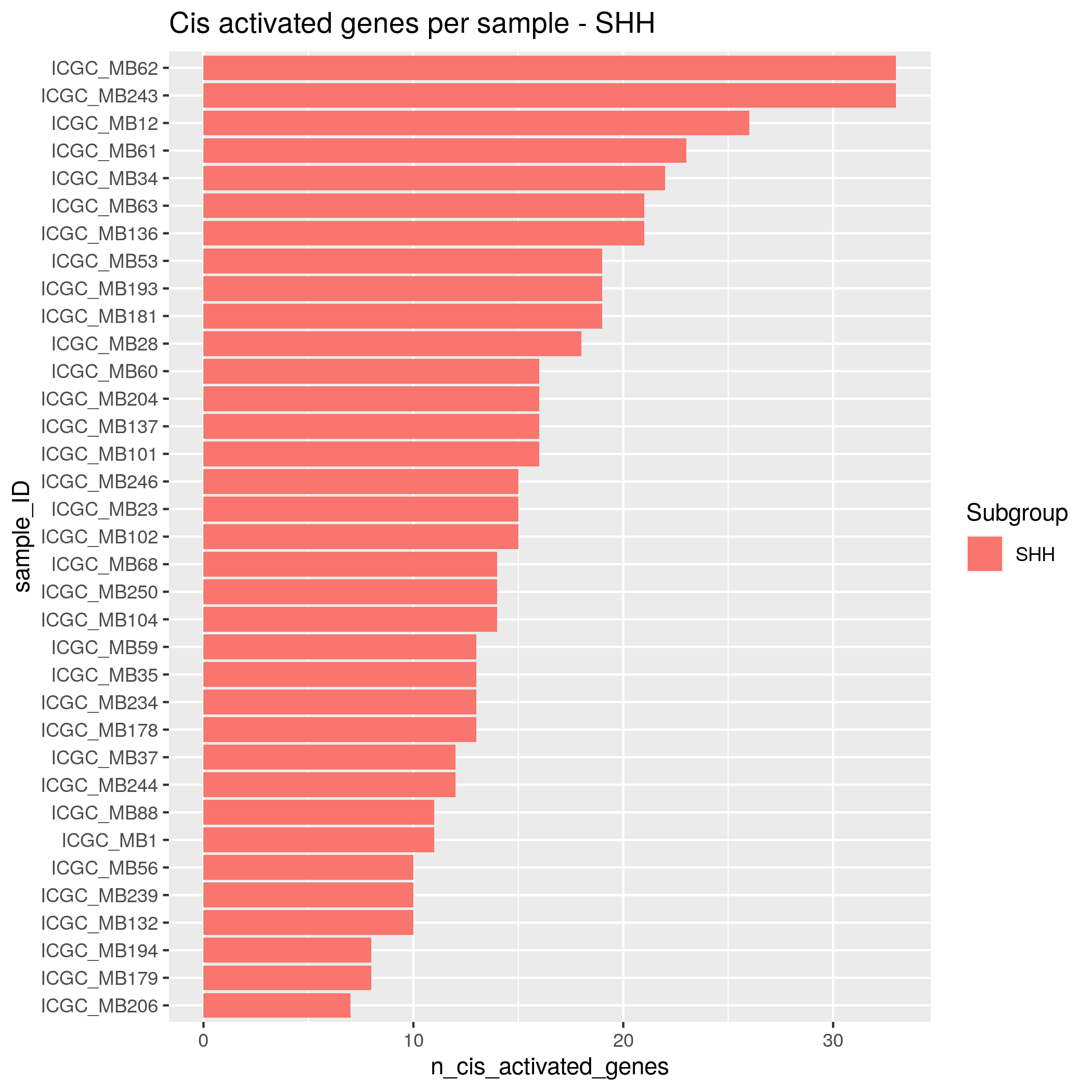
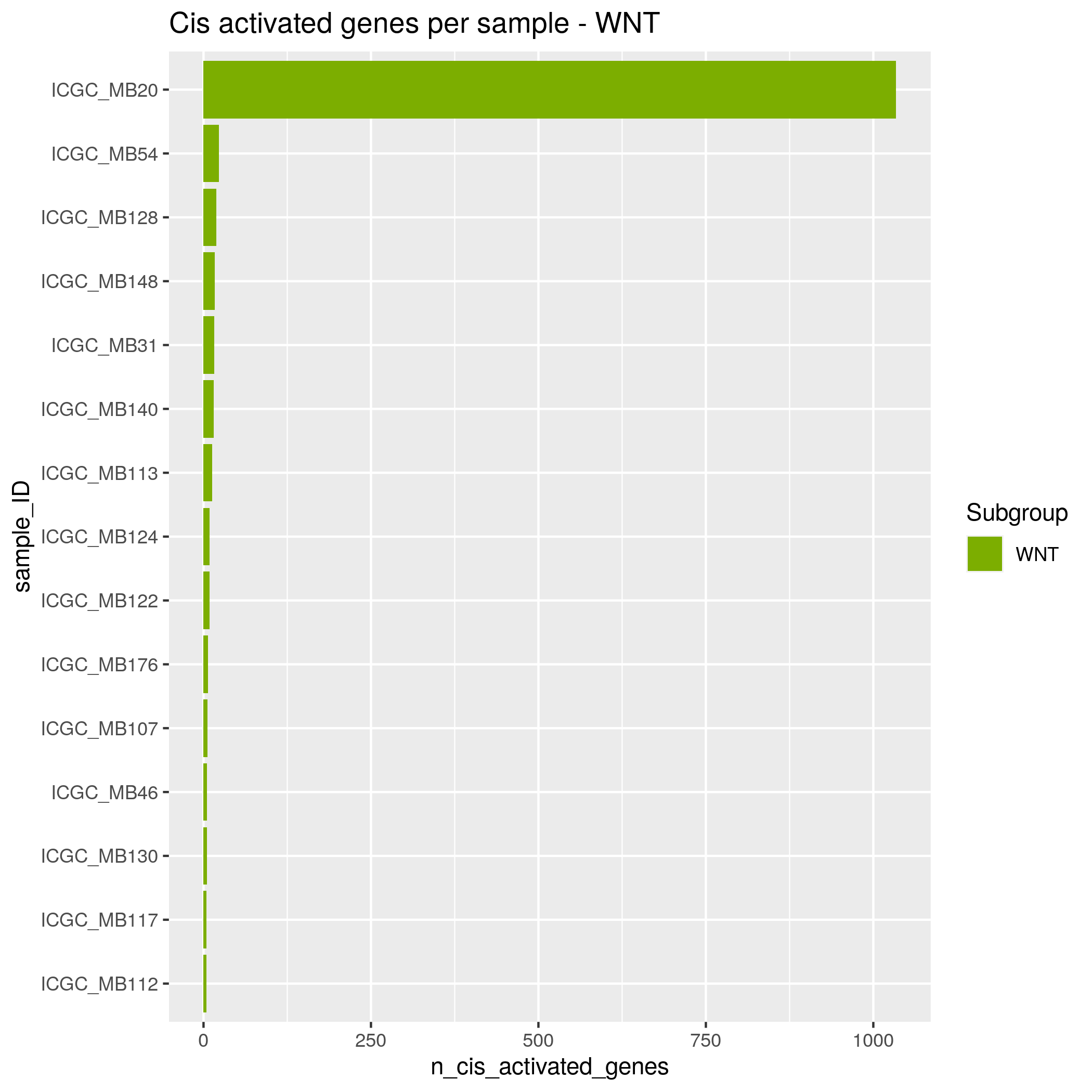

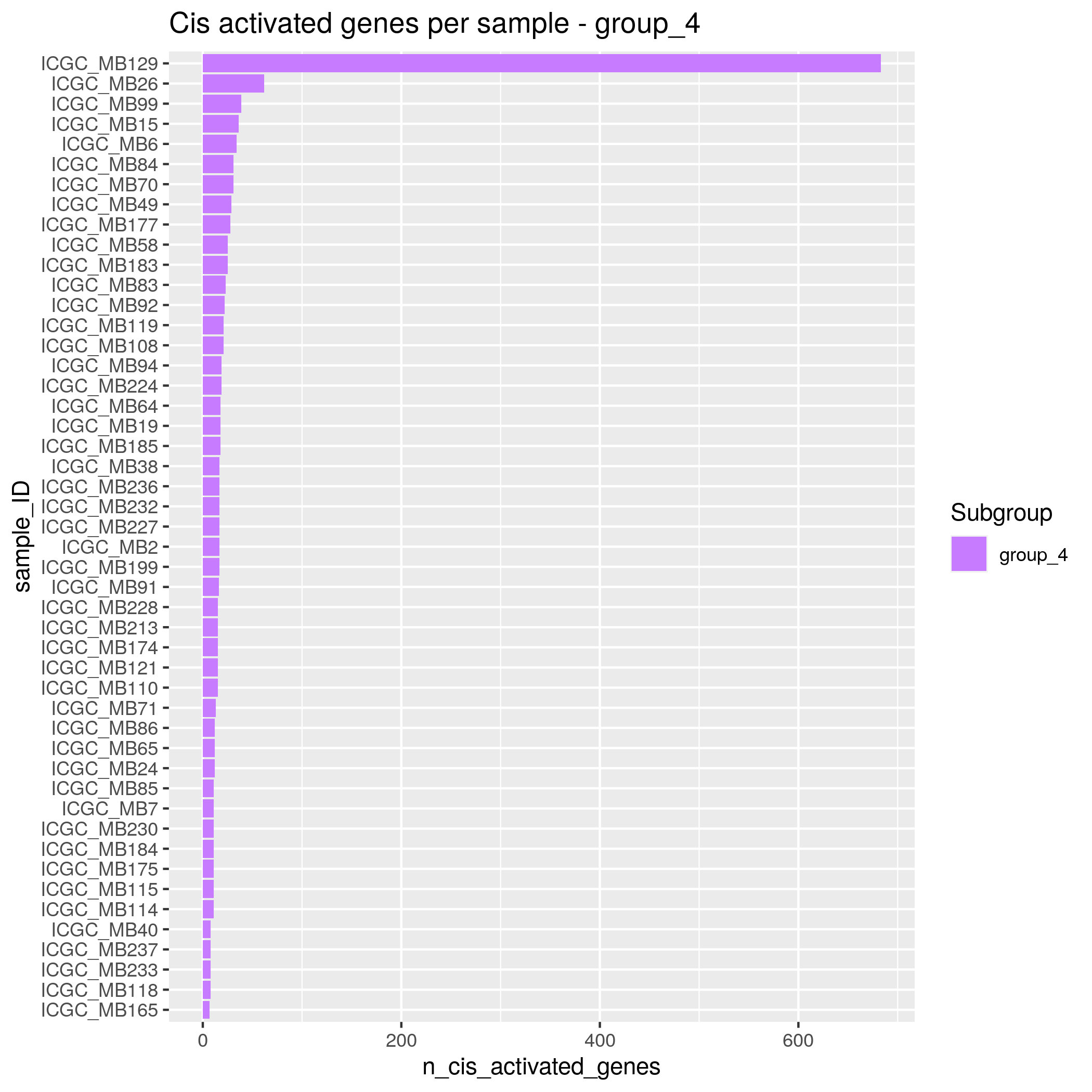
Algorithm insights Results
Detected Cis-Activated Samples Per Gene
Mutations highlighted
Input data
Applied Outlier High Expression
Mutations matched via
Sort by
Filter by
Filter by
Filter by
Detected Cis-Activated Samples Per Gene
SVs highlighted
Input data
Applied Outlier High Expression
Mutations matched via
Sort by
Filter by
Filter by
Filter by
Detected Cis-Activated Samples Per Gene
CNAs highlighted
Input data
Applied Outlier High Expression
Mutations matched via
Sort by
Filter by
Filter by
Filter by
Detected Cis-Activated Samples Per Gene
somatic SNVs highlighted
Input data
Applied Outlier High Expression
Mutations matched via
Sort by
Filter by
Filter by
Filter by
Detected Cis-Activated Samples Per Gene
somatic SNVs with relevant TF binding site highlighted
Input data
Applied Outlier High Expression
Mutations matched via
Sort by
Filter by
Filter by
Filter by
SVs: Recurrent SV TAD juxtaposition Results
| TADs | N of samples | Samples | N of samples with cis-activated genes affected | cis-activated genes |
|---|---|---|---|---|
| chr8:126800001-127840000 <-> chr8:128800001-129600000 | 3 | ICGC_MB198, ICGC_MB248, ICGC_MB50 | 2 | ICGC_MB198: PVT1; ICGC_MB248: PVT1 |
| chr8:127880001-128760000 <-> chr8:128800001-129600000 | 4 | ICGC_MB198, ICGC_MB248, ICGC_MB50, ICGC_MB94 | 2 | ICGC_MB198: PVT1, RP11-382A18.2; ICGC_MB248: PVT1 |
| chr12:11680001-12280000 <-> chr12:4920001-6160000 | 2 | ICGC_MB137, ICGC_MB243 | 1 | ICGC_MB137: RP11-1038A11.2 |
| chr12:4040001-4880000 <-> chr12:4920001-6160000 | 2 | ICGC_MB137, ICGC_MB34 | 1 | ICGC_MB137: RP11-1038A11.2 |
| chr12:4920001-6160000 <-> chr12:6200001-7240000 | 2 | ICGC_MB137, ICGC_MB243 | 1 | ICGC_MB137: RP11-1038A11.2 |
| chr17:71400001-72880000 <-> chr17:77760001-79080000 | 3 | ICGC_MB224, ICGC_MB243, ICGC_MB91 | 1 | ICGC_MB243: TBC1D16 |
| chr2:121040001-122600000 <-> chr2:123320001-124720000 | 2 | ICGC_MB250, ICGC_MB50 | 1 | ICGC_MB50: INHBB |
| chr2:121040001-122600000 <-> chr2:125400001-126400000 | 2 | ICGC_MB243, ICGC_MB250 | 1 | ICGC_MB243: CLASP1, CNTNAP5 |
| chr2:122640001-123280000 <-> chr2:124760001-125360000 | 2 | ICGC_MB243, ICGC_MB250 | 1 | ICGC_MB243: CNTNAP5 |
| chr2:13240001-15120000 <-> chr2:15800001-16800000 | 2 | ICGC_MB165, ICGC_MB183 | 1 | ICGC_MB183: AC092635.1 |
| chr2:15160001-15760000 <-> chr2:15800001-16800000 | 4 | ICGC_MB177, ICGC_MB183, ICGC_MB36, ICGC_MB6 | 1 | ICGC_MB36: AC008278.3 |
| chr2:15800001-16800000 <-> chr2:16840001-17920000 | 2 | ICGC_MB36, ICGC_MB6 | 1 | ICGC_MB36: AC008278.3 |
| chr2:222880001-223600000 <-> chr2:223640001-224760000 | 2 | ICGC_MB106, ICGC_MB250 | 1 | ICGC_MB250: ACSL3 |
| chr5:86760001-88040000 <-> chr5:88080001-89000000 | 2 | ICGC_MB19, ICGC_MB199 | 1 | ICGC_MB199: MEF2C |
| chr11:84400001-85320000 <-> chr20:54960001-56040000 | 2 | ICGC_MB65, ICGC_MB68 | 0 | |
| chr12:1840001-2920000 <-> chr12:29280001-30960000 | 2 | ICGC_MB137, ICGC_MB34 | 0 | |
| chr12:1840001-2920000 <-> chr12:31480001-32920000 | 2 | ICGC_MB137, ICGC_MB34 | 0 | |
| chr12:1840001-2920000 <-> chr12:4040001-4880000 | 2 | ICGC_MB137, ICGC_MB34 | 0 | |
| chr17:36480001-36880000 <-> chr2:15800001-16800000 | 2 | ICGC_MB137, ICGC_MB6 | 0 | |
| chr1:196000001-196760000 <-> chr1:196800001-197360000 | 2 | ICGC_MB65, ICGC_MB68 | 0 | |
| chr1:32640001-33600000 <-> chr1:57280001-59120000 | 2 | ICGC_MB65, ICGC_MB68 | 0 | |
| chr21:17120001-17880000 <-> chr21:17920001-18760000 | 2 | ICGC_MB19, ICGC_MB89 | 0 | |
| chr2:117840001-118560000 <-> chr2:123320001-124720000 | 2 | ICGC_MB102, ICGC_MB250 | 0 | |
| chr2:120200001-121000000 <-> chr2:129560001-130760000 | 2 | ICGC_MB243, ICGC_MB250 | 0 | |
| chr2:122640001-123280000 <-> chr2:123320001-124720000 | 2 | ICGC_MB102, ICGC_MB250 | 0 | |
| chr2:126440001-127760000 <-> chr2:128840001-129520000 | 2 | ICGC_MB243, ICGC_MB250 | 0 | |
| chr2:15160001-15760000 <-> chr2:16840001-17920000 | 4 | ICGC_MB165, ICGC_MB183, ICGC_MB36, ICGC_MB6 | 0 | |
| chr2:16840001-17920000 <-> chr2:17960001-18600000 | 2 | ICGC_MB165, ICGC_MB6 | 0 | |
| chr3:37000001-37360000 <-> chr3:38520001-39440000 | 2 | ICGC_MB1, ICGC_MB174 | 0 | |
| chr3:38040001-38480000 <-> chr3:38520001-39440000 | 2 | ICGC_MB131, ICGC_MB247 | 0 | |
| chr3:38520001-39440000 <-> chr3:39480001-40400000 | 2 | ICGC_MB247, ICGC_MB85 | 0 | |
| chr4:90080001-91240000 <-> chr4:92120001-93280000 | 2 | ICGC_MB110, ICGC_MB86 | 0 | |
| chr5:134240001-134800000 <-> chr5:134840001-135560000 | 2 | ICGC_MB121, ICGC_MB64 | 0 | |
| chr5:18440001-19600000 <-> chr5:19640001-20680000 | 2 | ICGC_MB137, ICGC_MB34 | 0 | |
| chr7:65200001-66760000 <-> chr7:71280001-72480000 | 3 | ICGC_MB65, ICGC_MB68, ICGC_MB71 | 0 | |
| chr8:128800001-129600000 <-> chr8:82720001-84080000 | 2 | ICGC_MB205, ICGC_MB89 | 0 | |
| chr8:82720001-84080000 <-> chr8:84120001-84840000 | 2 | ICGC_MB6, ICGC_MB89 | 0 | |
| chr9:19480001-20320000 <-> chr9:20360001-21200000 | 2 | ICGC_MB110, ICGC_MB83 | 0 |
Part III: Analysis of mutation types
Relation between mutations and cis activation Mutation Type
All
Overview
| no mutation | any mutation | |
|---|---|---|
| gene NOT cis activated | 4650473 | 2483495 |
| gene cis activated | 2450 | 1502 |
| cis activation frequency | 0.000526550729509171 | 0.000604427289047029 |
ODDS RATIO: 1.14798889139569
Chi-Square Test
Pearson's Chi-squared test with Yates' continuity correction
X-squared = 17.6132420723284, df = 1, p-value = 2.70696875780571e-05
Genehancer
| no mutation (via genehancer) | any mutation (via genehancer) | |
|---|---|---|
| gene NOT cis activated | 7030355 | 103613 |
| gene cis activated | 3761 | 191 |
| cis activation frequency | 0.000534679837523294 | 0.00184000616546569 |
ODDS RATIO: 3.44582359787565
Chi-Square Test
Pearson's Chi-squared test with Yates' continuity correction
X-squared = 312.627241863826, df = 1, p-value = 5.84662710554429e-70
Gene
| no mutation (via gene TAD) | any mutation (via gene TAD) | |
|---|---|---|
| gene NOT cis activated | 4658380 | 2475588 |
| gene cis activated | 2470 | 1482 |
| cis activation frequency | 0.000529946254438568 | 0.000598287492884739 |
ODDS RATIO: 1.12903601083864
Chi-Square Test
Pearson's Chi-squared test with Yates' continuity correction
X-squared = 13.5286629473679, df = 1, p-value = 0.000234947372916894
ChIP-Seq
| no mutation (with affected ChIP-Seq region) | any mutation (with affected ChIP-Seq region) | |
|---|---|---|
| gene NOT cis activated | 6931720 | 202248 |
| gene cis activated | 3749 | 203 |
| cis activation frequency | 0.000540554647421825 | 0.00100271176729184 |
ODDS RATIO: 1.85582650633447
Chi-Square Test
Pearson's Chi-squared test with Yates' continuity correction
X-squared = 75.0945270634014, df = 1, p-value = 4.48708413872907e-18
Relation between SVs and cis activation Mutation Type
All
Overview
| no SV | any SV | |
|---|---|---|
| gene NOT cis activated | 7060676 | 73292 |
| gene cis activated | 3862 | 90 |
| cis activation frequency | 0.000546674106643633 | 0.00122645880461149 |
ODDS RATIO: 2.24501863434368
Chi-Square Test
Pearson's Chi-squared test with Yates' continuity correction
X-squared = 59.4289815433565, df = 1, p-value = 1.26788049288852e-14
Genehancer
| no SV (via genehancer) | any SV (via genehancer) | |
|---|---|---|
| gene NOT cis activated | 7104174 | 29794 |
| gene cis activated | 3889 | 63 |
| cis activation frequency | 0.000547125145064134 | 0.00211005794286097 |
ODDS RATIO: 3.86266800954106
Chi-Square Test
Pearson's Chi-squared test with Yates' continuity correction
X-squared = 128.441434790913, df = 1, p-value = 8.98600075965905e-30
Gene
| no SV (via gene TAD) | any SV (via gene TAD) | |
|---|---|---|
| gene NOT cis activated | 7063396 | 70572 |
| gene cis activated | 3866 | 86 |
| cis activation frequency | 0.000547029387052581 | 0.00121713040278525 |
ODDS RATIO: 2.2264745292176
Chi-Square Test
Pearson's Chi-squared test with Yates' continuity correction
X-squared = 55.5651909586256, df = 1, p-value = 9.04104638855294e-14
ChIP-Seq
| no SV (with affected ChIP-Seq region) | any SV (with affected ChIP-Seq region) | |
|---|---|---|
| gene NOT cis activated | 7079354 | 54614 |
| gene cis activated | 3872 | 80 |
| cis activation frequency | 0.00054664357737562 | 0.00146268329250009 |
ODDS RATIO: 2.67820787029149
Chi-Square Test
Pearson's Chi-squared test with Yates' continuity correction
X-squared = 80.6572839409651, df = 1, p-value = 2.68463131145813e-19
Relation between CNAs and cis activation Mutation Type
All
Overview
| no CNA | any CNA | |
|---|---|---|
| gene NOT cis activated | 6991542 | 142426 |
| gene cis activated | 3783 | 169 |
| cis activation frequency | 0.000540789741720363 | 0.0011851747957502 |
ODDS RATIO: 2.19297689912693
Chi-Square Test
Pearson's Chi-squared test with Yates' continuity correction
X-squared = 103.702796066654, df = 1, p-value = 2.35059261729545e-24
Genehancer
| no CNA (via genehancer) | any CNA (via genehancer) | |
|---|---|---|
| gene NOT cis activated | 7081007 | 52961 |
| gene cis activated | 3830 | 122 |
| cis activation frequency | 0.000540591124397075 | 0.00229828758736319 |
ODDS RATIO: 4.25892413171858
Chi-Square Test
Pearson's Chi-squared test with Yates' continuity correction
X-squared = 291.000982510995, df = 1, p-value = 3.00921044587203e-65
Gene
| no CNA (via gene TAD) | any CNA (via gene TAD) | |
|---|---|---|
| gene NOT cis activated | 6997006 | 136962 |
| gene cis activated | 3802 | 150 |
| cis activation frequency | 0.000543080170174643 | 0.00109399614913355 |
ODDS RATIO: 2.01553945306931
Chi-Square Test
Pearson's Chi-squared test with Yates' continuity correction
X-squared = 72.767267652562, df = 1, p-value = 1.45876293284383e-17
ChIP-Seq
| no CNA (with affected ChIP-Seq region) | any CNA (with affected ChIP-Seq region) | |
|---|---|---|
| gene NOT cis activated | 7022824 | 111144 |
| gene cis activated | 3835 | 117 |
| cis activation frequency | 0.000545778584103768 | 0.00105158141666891 |
ODDS RATIO: 1.92773021897383
Chi-Square Test
Pearson's Chi-squared test with Yates' continuity correction
X-squared = 49.728223186276, df = 1, p-value = 1.7658734518754e-12
Relation between somatic SNVs and cis activation Mutation Type
All
Overview
| no somatic_SNV | any somatic_SNV | |
|---|---|---|
| gene NOT cis activated | 4744141 | 2389827 |
| gene cis activated | 2531 | 1421 |
| cis activation frequency | 0.000533215693016075 | 0.000594250366335905 |
ODDS RATIO: 1.11453332218632
Chi-Square Test
Pearson's Chi-squared test with Yates' continuity correction
X-squared = 10.5950837036705, df = 1, p-value = 0.00113388765565416
Genehancer
| no somatic_SNV (via genehancer) | any somatic_SNV (via genehancer) | |
|---|---|---|
| gene NOT cis activated | 7099238 | 34730 |
| gene cis activated | 3896 | 56 |
| cis activation frequency | 0.000548490286118775 | 0.00160984304030357 |
ODDS RATIO: 2.93816398843292
Chi-Square Test
Pearson's Chi-squared test with Yates' continuity correction
X-squared = 68.5639381718932, df = 1, p-value = 1.22829844173787e-16
Gene
| no somatic_SNV (via gene TAD) | any somatic_SNV (via gene TAD) | |
|---|---|---|
| gene NOT cis activated | 4747175 | 2386793 |
| gene cis activated | 2538 | 1414 |
| cis activation frequency | 0.000534348075346868 | 0.000592075980013458 |
ODDS RATIO: 1.10809827317813
Chi-Square Test
Pearson's Chi-squared test with Yates' continuity correction
X-squared = 9.46645840442032, df = 1, p-value = 0.00209262994962569
ChIP-Seq
| no somatic_SNV (with affected ChIP-Seq region) | any somatic_SNV (with affected ChIP-Seq region) | |
|---|---|---|
| gene NOT cis activated | 7071487 | 62481 |
| gene cis activated | 3890 | 62 |
| cis activation frequency | 0.000534348075346868 | 0.000592075980013458 |
ODDS RATIO: 1.80386845415916
Chi-Square Test
Pearson's Chi-squared test with Yates' continuity correction
X-squared = 21.0497764564183, df = 1, p-value = 4.47505058614993e-06
Relation between "relevant" somatic SNVs and cis activation Mutation Type
All
Overview
| no relevant somatic SNV | any relevant somatic SNV | |
|---|---|---|
| gene NOT cis activated | 4983903 | 2150065 |
| gene cis activated | 2671 | 1281 |
| cis activation frequency | 0.000535638295952291 | 0.000595441179614994 |
ODDS RATIO: 1.11171440584111
Chi-Square Test
Pearson's Chi-squared test with Yates' continuity correction
X-squared = 9.60583094458175, df = 1, p-value = 0.00193960488908808
Genehancer
| no relevant somatic SNV (via genehancer) | any relevant somatic SNV (via genehancer) | |
|---|---|---|
| gene NOT cis activated | 7102917 | 31051 |
| gene cis activated | 3901 | 51 |
| cis activation frequency | 0.000548909511964426 | 0.00163976593145135 |
ODDS RATIO: 2.99057965440769
Chi-Square Test
Pearson's Chi-squared test with Yates' continuity correction
X-squared = 64.6352771894726, df = 1, p-value = 9.01272182482257e-16
TF binding site analysis ROC
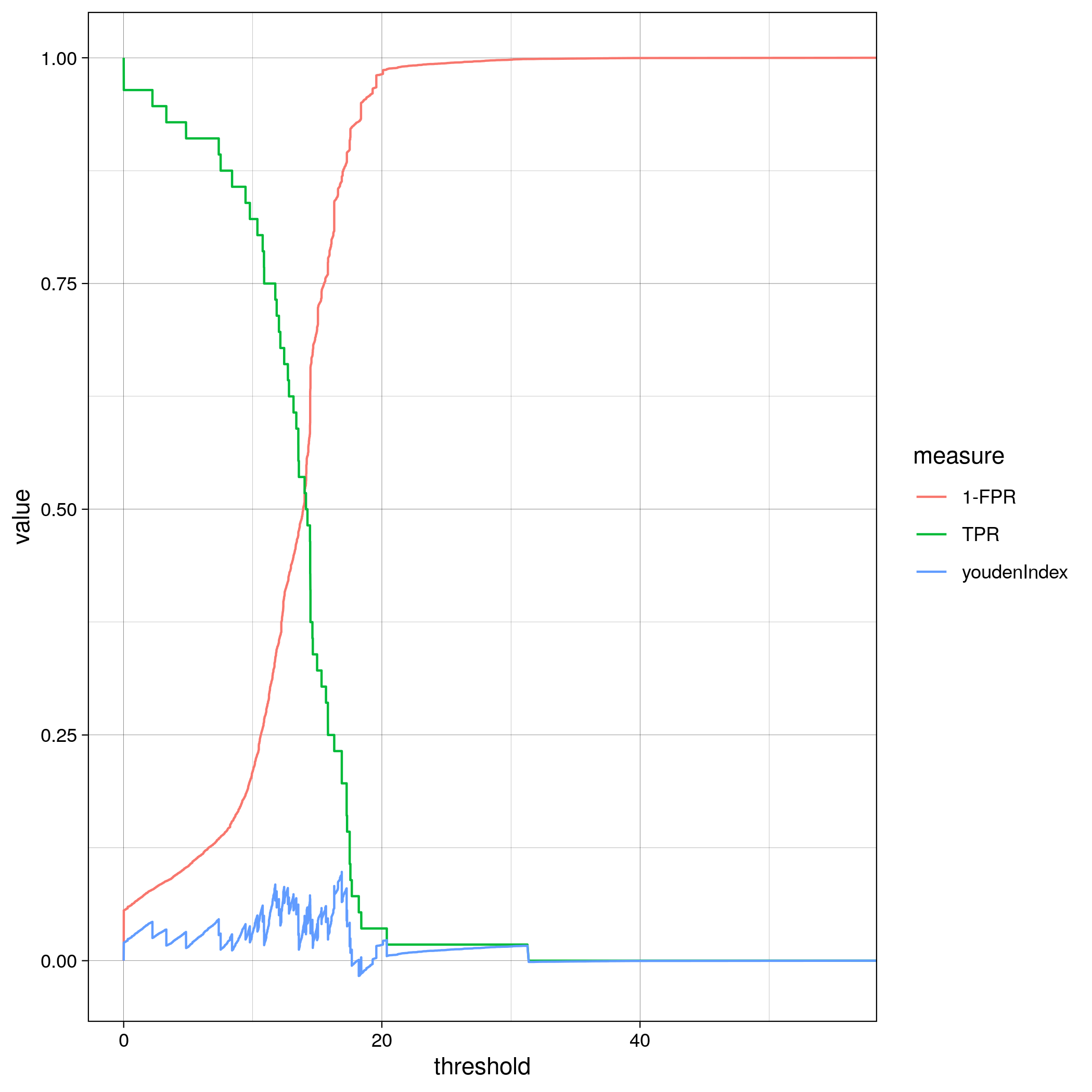
Gene
| no relevant somatic SNV (via gene TAD) | any relevant somatic SNV (via gene TAD) | |
|---|---|---|
| gene NOT cis activated | 4986639 | 2147329 |
| gene cis activated | 2678 | 1274 |
| cis activation frequency | 0.000536746813241171 | 0.000592943414860726 |
ODDS RATIO: 1.10476064581419
Chi-Square Test
Pearson's Chi-squared test with Yates' continuity correction
X-squared = 8.46993179365657, df = 1, p-value = 0.00361064969537433
TF binding site analysis ROC
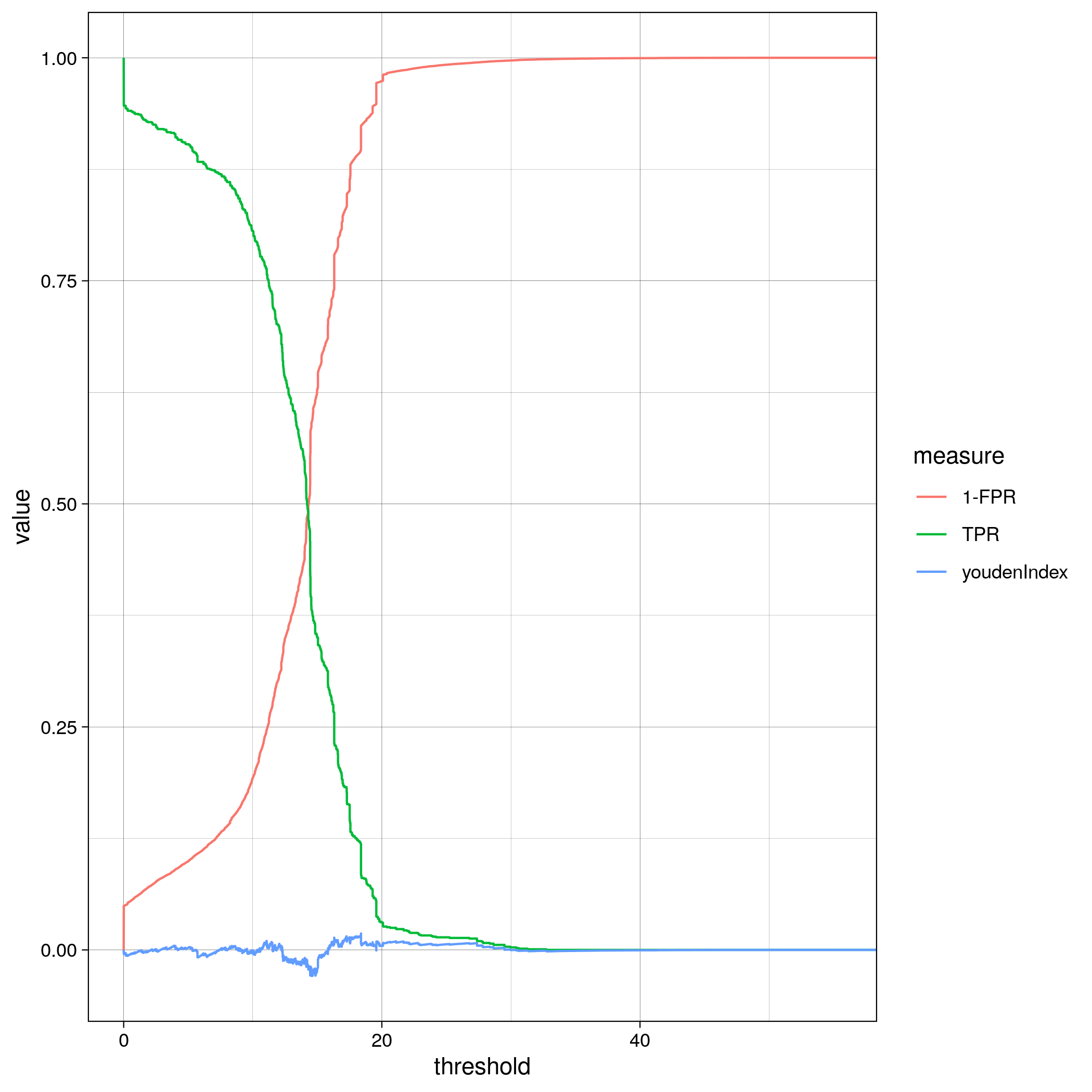
ChIP-Seq
| no relevant somatic SNV (with affected ChIP-Seq region) | any relevant somatic SNV (with affected ChIP-Seq region) | |
|---|---|---|
| gene NOT cis activated | 7079927 | 54041 |
| gene cis activated | 3900 | 52 |
| cis activation frequency | 0.000536746813241171 | 0.000592943414860726 |
ODDS RATIO: 1.74680384646225
Chi-Square Test
Pearson's Chi-squared test with Yates' continuity correction
X-squared = 15.6344155837162, df = 1, p-value = 7.68432345259015e-05
TF binding site analysis ROC
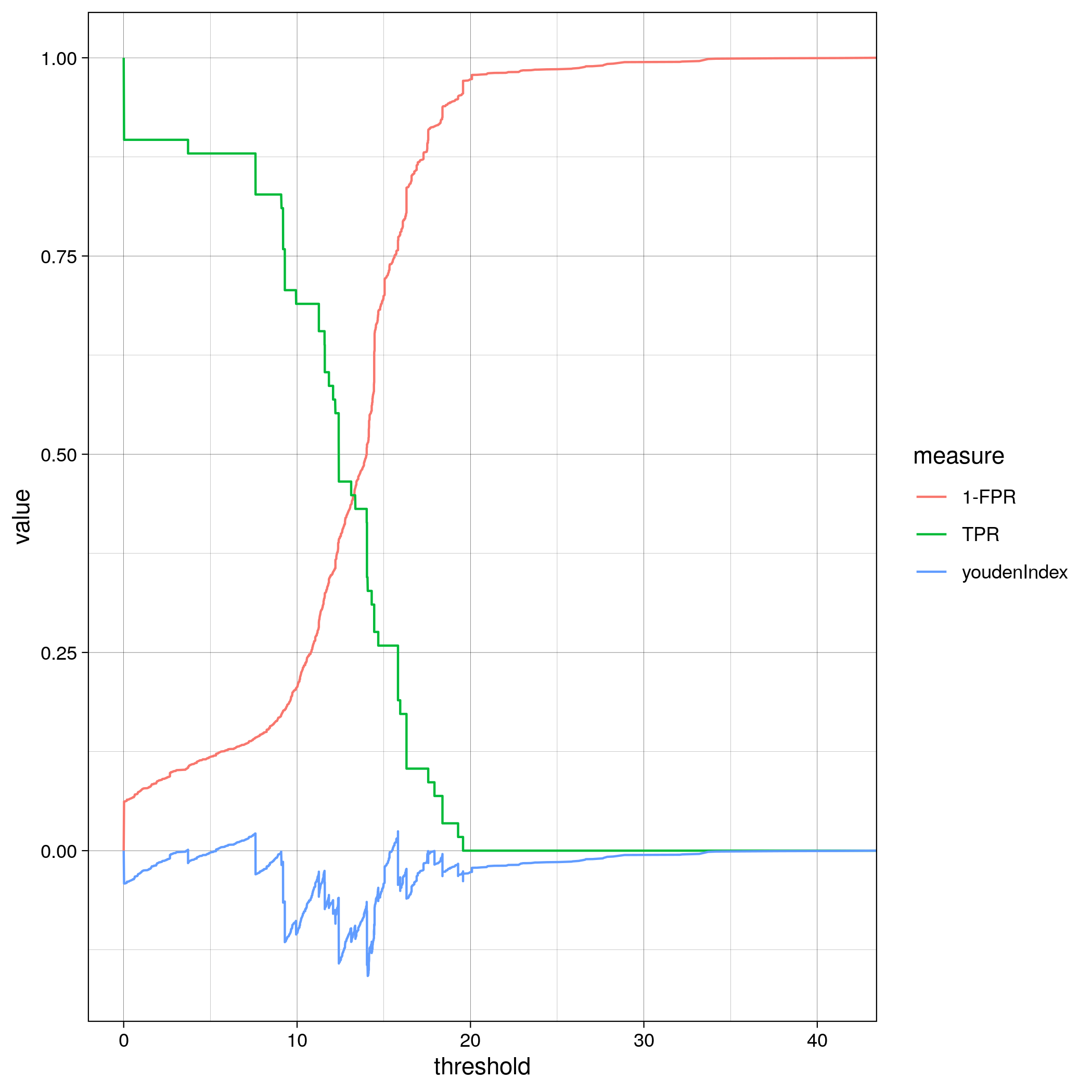
Part IV: Analysis by gene
GFI1B
protein_codingchr9 : 135 820 932 - 135 867 083
4 cis-activated samples 4 cis-activated samples with SVs 3 cis-activated samples with CNAs 2 cis-activated samples with somatic SNVsPRDM6
protein_codingchr5 : 122 424 816 - 122 529 960
7 cis-activated samples 7 cis-activated samples with SVs 7 cis-activated samples with CNAs 6 cis-activated samples with somatic SNVsPVT1
processed_transcriptchr8 : 128 806 779 - 129 113 499
2 cis-activated samples 2 cis-activated samples with SVs 2 cis-activated samples with CNAs 2 cis-activated samples with somatic SNVsMYC
protein_codingchr8 : 128 747 680 - 128 753 674
1 cis-activated samples 1 cis-activated samples with SVs 0 cis-activated samples with CNAs 0 cis-activated samples with somatic SNVs